19. One-Way ANOVA
One-way Analysis of Variance (ANOVA) is a multiple comparison technique testing whether the means of 3 or more groups are different. The hypotheses are as follows.
\(H_0\) (null): There is no difference between the means; \(\mu_1 = \mu_2 = \ldots = \mu_n\)
\(H_a\) (alternative): There is at least one group mean that differs from the overall mean.
ANOVA uses the F-test for testing statistical significance, which measures the ratio of the variance between groups to the variance within groups.
When there are only 2 groups, a t-test may be used instead. However, with comparing 3 or more groups, ANOVA should be used since it corrects for the family-wise error rate FWER holding the probability of a Type I error steady no matter the number of comparisons.
In this notebook, let’s see how a 1-way ANOVA relates to regression problems.
19.1. Data
Let’s generate 50 samples from 3 different normal distributions. These normal distributions are defined as follows.
\(A \sim \mathcal{N} (0, 1)\)
\(B \sim \mathcal{N} (1, 1)\)
\(C \sim \mathcal{N} (1.5, 1)\)
These groups are often called treatments.
[1]:
import numpy as np
import pandas as pd
np.random.seed(37)
n = 50
df = pd.concat([
pd.DataFrame({'group': 'A', 'y': pd.Series(np.random.normal(0, 1, n))}),
pd.DataFrame({'group': 'B', 'y': pd.Series(np.random.normal(1, 1, n))}),
pd.DataFrame({'group': 'C', 'y': pd.Series(np.random.normal(1.5, 1, n))})
])
[2]:
df.head()
[2]:
| group | y | |
|---|---|---|
| 0 | A | -0.054464 |
| 1 | A | 0.674308 |
| 2 | A | 0.346647 |
| 3 | A | -1.300346 |
| 4 | A | 1.518512 |
19.2. Means
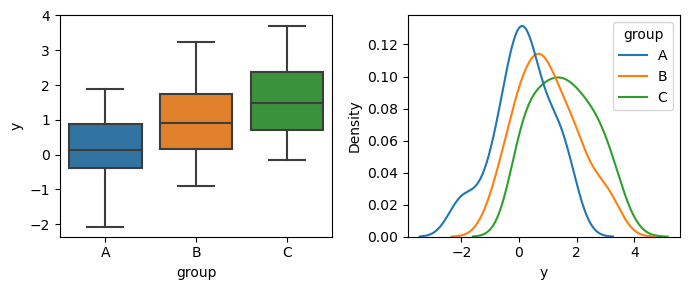
You can see that the empirical means and standard deviations are close to the real ones, but not exactly.
[3]:
df.groupby(['group']).agg(['mean', 'std'])
[3]:
| y | ||
|---|---|---|
| mean | std | |
| group | ||
| A | 0.139065 | 0.998983 |
| B | 0.982454 | 1.057929 |
| C | 1.552792 | 1.052197 |
[4]:
import matplotlib.pyplot as plt
import seaborn as sns
fig, ax = plt.subplots(1, 2, figsize=(7, 3))
sns.boxplot(df, x='group', y='y', ax=ax[0])
sns.kdeplot(df, hue='group', x='y', ax=ax[1])
fig.tight_layout()
19.3. t-test
Let’s see what happens when we conduct all pairwise t-tests. You can see that group A is very different from B and C, but groups B and C are comparatively close.
[5]:
from scipy import stats
from scipy import special
import itertools
def do_test(pair):
x, y = pair
x = df[df['group']==x]['y']
y = df[df['group']==y]['y']
t = stats.ttest_ind(x, y)
s = t.statistic
p = t.pvalue
a = 0.05 / special.comb(3, 2) # bonferroni's correction, however, losing power
return {
'x': pair[0],
'y': pair[1],
'statistic': s,
'pvalue': p,
'alpha': a,
'is_sig': p < a
}
t_test = itertools.combinations(df['group'].unique(), 2)
t_test = map(lambda tup: do_test(tup), t_test)
t_test = pd.DataFrame(t_test)
t_test
[5]:
| x | y | statistic | pvalue | alpha | is_sig | |
|---|---|---|---|---|---|---|
| 0 | A | B | -4.098587 | 8.576614e-05 | 0.016667 | True |
| 1 | A | C | -6.889943 | 5.417835e-10 | 0.016667 | True |
| 2 | B | C | -2.702849 | 8.104302e-03 | 0.016667 | True |
19.4. 1-way ANOVA
Now we will apply a 1-way ANOVA. The report F statistic and p-value are shown. The test would lead us to reject the null hypothesis.
[6]:
F, p = stats.f_oneway(*[df[df['group']==g]['y'] for g in ['A', 'B', 'C']])
[7]:
F
[7]:
23.533902104682596
[8]:
p
[8]:
1.3590719185638985e-09
19.5. Tukey’s HSD
Here is a post-hoc test using Tukey’s Honestly Significant Difference test. This test will see which two pair of groups are different.
[9]:
hsd = stats.tukey_hsd(*[df[df['group']==g]['y'] for g in ['A', 'B', 'C']])
[10]:
hsd.statistic
[10]:
array([[ 0. , -0.84338966, -1.41372695],
[ 0.84338966, 0. , -0.57033729],
[ 1.41372695, 0.57033729, 0. ]])
[11]:
hsd.pvalue
[11]:
array([[1.00000000e+00, 2.26787268e-04, 6.66437017e-10],
[2.26787268e-04, 1.00000000e+00, 1.82679298e-02],
[6.66437017e-10, 1.82679298e-02, 1.00000000e+00]])
[12]:
hsd._nobs
[12]:
3
[13]:
hsd._ntreatments
[13]:
150
[14]:
hsd._stand_err
[14]:
0.14661287403767
19.6. Xy, means
Before we move onto regression, keep in mind the group means and overall mean.
[15]:
df.groupby(['group']).agg(['mean', 'std'])
[15]:
| y | ||
|---|---|---|
| mean | std | |
| group | ||
| A | 0.139065 | 0.998983 |
| B | 0.982454 | 1.057929 |
| C | 1.552792 | 1.052197 |
[16]:
df['y'].mean()
[16]:
0.8914368828670105
19.7. Regression
Here, we regress y ~ g but with g dummy-encoded (one-hot encoded).
[17]:
X = df.drop(columns=['y'])
y = df['y']
[18]:
from sklearn.preprocessing import OneHotEncoder
ohe = OneHotEncoder()
ohe.fit(X)
[18]:
OneHotEncoder()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
OneHotEncoder()
[19]:
X = pd.DataFrame(
ohe.transform(X).todense(),
columns=ohe.get_feature_names_out()
)
X
[19]:
| group_A | group_B | group_C | |
|---|---|---|---|
| 0 | 1.0 | 0.0 | 0.0 |
| 1 | 1.0 | 0.0 | 0.0 |
| 2 | 1.0 | 0.0 | 0.0 |
| 3 | 1.0 | 0.0 | 0.0 |
| 4 | 1.0 | 0.0 | 0.0 |
| ... | ... | ... | ... |
| 145 | 0.0 | 0.0 | 1.0 |
| 146 | 0.0 | 0.0 | 1.0 |
| 147 | 0.0 | 0.0 | 1.0 |
| 148 | 0.0 | 0.0 | 1.0 |
| 149 | 0.0 | 0.0 | 1.0 |
150 rows × 3 columns
[20]:
from sklearn.linear_model import LinearRegression
m = LinearRegression()
m.fit(X.drop(columns=['group_A']), y)
[20]:
LinearRegression()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
LinearRegression()
[21]:
m.intercept_, m.coef_
[21]:
(0.13906468043625642, array([0.84338966, 1.41372695]))
[22]:
m.coef_
[22]:
array([0.84338966, 1.41372695])
19.8. Means vs coefficients
If you notice, the intercept of the regression model is the mean of A. If you add the coefficients to the intercept, you recover the means of B and C!
[23]:
df.groupby(['group']).mean()
[23]:
| y | |
|---|---|
| group | |
| A | 0.139065 |
| B | 0.982454 |
| C | 1.552792 |
[24]:
m.intercept_, m.coef_ + m.intercept_
[24]:
(0.13906468043625642, array([0.98245434, 1.55279163]))
19.9. Test statistics vs coefficients
The coefficients are the test statistics Tukey’s HSD!
[25]:
hsd.statistic
[25]:
array([[ 0. , -0.84338966, -1.41372695],
[ 0.84338966, 0. , -0.57033729],
[ 1.41372695, 0.57033729, 0. ]])
[26]:
m.coef_
[26]:
array([0.84338966, 1.41372695])
When using Tukey’s HSD, the p-values are given.
[27]:
hsd.pvalue
[27]:
array([[1.00000000e+00, 2.26787268e-04, 6.66437017e-10],
[2.26787268e-04, 1.00000000e+00, 1.82679298e-02],
[6.66437017e-10, 1.82679298e-02, 1.00000000e+00]])
We can also compute the p-values as below. The main complication is with computing the standard error SE.
[28]:
def get_se(df):
'''
Computes standard error.
Adapted from https://github.com/scipy/scipy/blob/v1.10.1/scipy/stats/_hypotests.py.
When groups are unequal, this code should be adjusted too.
'''
_v = np.array([np.var(df[df['group']==g]['y'], ddof=1) for g in df['group'].unique()])
_s = np.array([df[df['group']==g].shape[0] - 1 for g in df['group'].unique()])
_n = df.shape[0] - df['group'].unique().shape[0]
_mse = np.sum(_v * _s) / _n
_norm = 2 / df[df['group']=='A'].shape[0]
_se = np.sqrt(_norm * _mse / 2)
return _se
def get_pvalue(m_0, m_1, se, k, dof):
diff = np.abs(m_0 - m_1)
t = diff / se
print(f'{t=}, {k=}, {dof=}')
return stats.distributions.studentized_range.sf(t, k, dof)
[29]:
_mean = df.groupby(['group']).mean()['y'].to_numpy()
_se = get_se(df)
_k = df['group'].unique().shape[0]
_dof = df.shape[0] - _k
[30]:
_mean
[30]:
array([0.13906468, 0.98245434, 1.55279163])
[31]:
_se
[31]:
0.14661287403767
[32]:
_k
[32]:
3
[33]:
_dof
[33]:
147
[34]:
get_pvalue(_mean[0], _mean[1], _se, _k, _dof)
t=5.752493850354061, k=3, dof=147
[34]:
0.00022678726812552785
[35]:
get_pvalue(_mean[0], _mean[2], _se, _k, _dof)
t=9.64258398375435, k=3, dof=147
[35]:
6.664370166831191e-10
[36]:
get_pvalue(_mean[1], _mean[2], _se, _k, _dof)
t=3.8900901334002893, k=3, dof=147
[36]:
0.01826792981617653
Here’s yet another way to compute the p-values.
[37]:
1 - stats.studentized_range.cdf(np.abs(_mean[0] - _mean[1]) / _se, _k, _dof)
[37]:
0.00022678726812552785
[38]:
1 - stats.studentized_range.cdf(np.abs(_mean[0] - _mean[2]) / _se, _k, _dof)
[38]:
6.664370166831191e-10
[39]:
1 - stats.studentized_range.cdf(np.abs(_mean[1] - _mean[2]) / _se, _k, _dof)
[39]:
0.01826792981617653