3. Principal Component Analysis - Iris
Let’s take a look at PCA again using the Iris dataset as an example to reinforce some concepts.
3.1. Load data
[1]:
from sklearn.datasets import load_iris
X, y = load_iris(return_X_y=True, as_frame=True)
X.columns = ['s_length', 's_width', 'p_length', 'p_width']
X.shape, y.shape
[1]:
((150, 4), (150,))
[2]:
X.head()
[2]:
| s_length | s_width | p_length | p_width | |
|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 |
3.2. PCA
The procedure of PCA to work is to
map
Xto z-scores,Z,compute the covariance matrix
CofZ, andapply singular-value decomposition (SVD) to
Zresulting inU,S,VT.
Here are the denotations.
X: dataZ: data mapped to z-scoresC: covariance matrix ofZU: left-hand eigen-vectorsS: eigen-valuesVT: right-hand eigen-vectors
Getting Z and C is performed as follows.
[3]:
from scipy.stats import zscore
Z = X.apply(zscore)
C = Z.cov()
SVD is applied to C as follows.
[4]:
from numpy.linalg import svd
U, S, VT = svd(C, full_matrices=False)
U.shape, S.shape, VT.shape
[4]:
((4, 4), (4,), (4, 4))
[5]:
U
[5]:
array([[-0.52106591, -0.37741762, 0.71956635, 0.26128628],
[ 0.26934744, -0.92329566, -0.24438178, -0.12350962],
[-0.5804131 , -0.02449161, -0.14212637, -0.80144925],
[-0.56485654, -0.06694199, -0.63427274, 0.52359713]])
[6]:
S
[6]:
array([2.93808505, 0.9201649 , 0.14774182, 0.02085386])
[7]:
VT
[7]:
array([[-0.52106591, 0.26934744, -0.5804131 , -0.56485654],
[-0.37741762, -0.92329566, -0.02449161, -0.06694199],
[ 0.71956635, -0.24438178, -0.14212637, -0.63427274],
[ 0.26128628, -0.12350962, -0.80144925, 0.52359713]])
3.3. Interpretation of S
When S is normalized so that the elements sum to unit, each resulting element is the amount of variance accounted for by its corresponding principal component. According to what we see below, the first 2 principal components account for (0.73 + 0.22) 95% of the variance in the data. Note that the elements in S are produced from largest to smallest; the first principal component accounts for the largest variance, followed by the second and so on.
[8]:
S / S.sum()
[8]:
array([0.72962445, 0.22850762, 0.03668922, 0.00517871])
3.4. Interpretation of U and VT
U and VT are just transpose of one another and they represent the strength of association between the features in X and principal components.
For
U, the columns correspond to principal components and rows to features.For
VT, the opposite is true; columns correspond to features and rows to principal components.
Each element in U or VT gives you a strength of association between the corresponding feature and principal component.
[9]:
import pandas as pd
pd.DataFrame(U, columns=X.columns, index=[f'PC-{i}' for i in range(U.shape[1])])
[9]:
| s_length | s_width | p_length | p_width | |
|---|---|---|---|---|
| PC-0 | -0.521066 | -0.377418 | 0.719566 | 0.261286 |
| PC-1 | 0.269347 | -0.923296 | -0.244382 | -0.123510 |
| PC-2 | -0.580413 | -0.024492 | -0.142126 | -0.801449 |
| PC-3 | -0.564857 | -0.066942 | -0.634273 | 0.523597 |
[10]:
pd.DataFrame(VT, columns=[f'PC-{i}' for i in range(VT.shape[1])], index=X.columns)
[10]:
| PC-0 | PC-1 | PC-2 | PC-3 | |
|---|---|---|---|---|
| s_length | -0.521066 | 0.269347 | -0.580413 | -0.564857 |
| s_width | -0.377418 | -0.923296 | -0.024492 | -0.066942 |
| p_length | 0.719566 | -0.244382 | -0.142126 | -0.634273 |
| p_width | 0.261286 | -0.123510 | -0.801449 | 0.523597 |
3.5. Loadings squared
The loadings gives you the Pearson correlation between a feature and PC, while the the loadings squared gives you how much of a feature’s variance is explained by each principal component. The sum of the variance explained by the principal components (for a feature) should sum to one.
Loadings L and loadings squared I is computed as follows.
\(L = U \times \sqrt{S}\)
\(I = L^2\)
[11]:
import numpy as np
L = U * np.sqrt(S)
pd.DataFrame(L, columns=[f'PC-{i}' for i in range(L.shape[1])], index=X.columns)
[11]:
| PC-0 | PC-1 | PC-2 | PC-3 | |
|---|---|---|---|---|
| s_length | -0.893151 | -0.362039 | 0.276581 | 0.037732 |
| s_width | 0.461684 | -0.885673 | -0.093934 | -0.017836 |
| p_length | -0.994877 | -0.023494 | -0.054629 | -0.115736 |
| p_width | -0.968212 | -0.064214 | -0.243797 | 0.075612 |
From the table above, ignoring the sign (positive or negative), for s_length,
it is most correlated to the first PC,
followed by the second,
the third, and
very little to the fourth.
In contrast, note how s_width is most correlated to the second PC.
[12]:
import numpy as np
I = np.power(L, 2)
pd.DataFrame(I, columns=[f'PC-{i}' for i in range(I.shape[1])], index=X.columns)
[12]:
| PC-0 | PC-1 | PC-2 | PC-3 | |
|---|---|---|---|---|
| s_length | 0.797719 | 0.131072 | 0.076497 | 0.001424 |
| s_width | 0.213152 | 0.784417 | 0.008824 | 0.000318 |
| p_length | 0.989780 | 0.000552 | 0.002984 | 0.013395 |
| p_width | 0.937434 | 0.004123 | 0.059437 | 0.005717 |
From the table above, for s_length,
80% of its variance is explained by the first PC,
13% by the second,
8% by the third, and
none by the fourth.
On the contrary, note how most of the variance of s_width is captured by the second PC.
3.6. Projection into PC space
PCA is typically used to reduce dimensionality, or, in other words, represent the data in lower dimension. Once you get U or VT, you can project the z-scaled data Z to PC space by a simple multiplication. From the looks of things above, it seems the first PCs account 95% of the data’s variance, and so we can project the 4-dimensional Z data to 2-dimensional PC data/representation.
[13]:
P = pd.DataFrame(np.dot(Z, U[:,:2]), columns=['PC-0', 'PC-1'])
Z.shape, P.shape
[13]:
((150, 4), (150, 2))
[14]:
P.head()
[14]:
| PC-0 | PC-1 | |
|---|---|---|
| 0 | 2.264703 | -0.480027 |
| 1 | 2.080961 | 0.674134 |
| 2 | 2.364229 | 0.341908 |
| 3 | 2.299384 | 0.597395 |
| 4 | 2.389842 | -0.646835 |
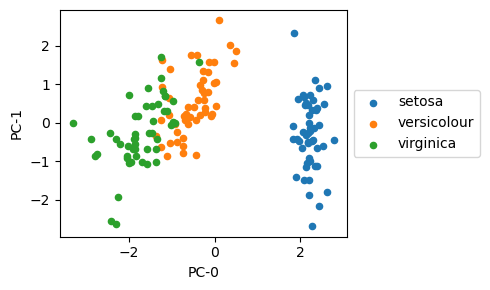
From the scatter plot below, the different iris flower species seem to be linearly separable in 2D PC space.
[15]:
import matplotlib.pyplot as plt
import seaborn as sns
fig, ax = plt.subplots(figsize=(5, 3))
for label, color, clazz in zip(y.unique(), sns.color_palette('tab10'), ['setosa', 'versicolour', 'virginica']):
P[y==label].plot(kind='scatter', x='PC-0', y='PC-1', color=color, ax=ax, label=clazz)
ax.legend(loc='center left', bbox_to_anchor=(1, 0.5))
fig.tight_layout()
3.7. Another view of U and VT
If we regress each PC on Z, we recover the eigenvectors.
\(P_0 \sim Z\)
\(P_1 \sim Z\)
\(P_2 \sim Z\)
\(P_3 \sim Z\)
Compare the 2 regressions below to VT.
[16]:
from sklearn.linear_model import LinearRegression
m = LinearRegression(fit_intercept=False)
m.fit(Z, P['PC-0'])
m.coef_
[16]:
array([-0.52106591, 0.26934744, -0.5804131 , -0.56485654])
[17]:
m = LinearRegression(fit_intercept=False)
m.fit(Z, P['PC-1'])
m.coef_
[17]:
array([-0.37741762, -0.92329566, -0.02449161, -0.06694199])
[18]:
pd.DataFrame(VT, columns=X.columns, index=[f'PC-{i}' for i in range(VT.shape[1])])
[18]:
| s_length | s_width | p_length | p_width | |
|---|---|---|---|---|
| PC-0 | -0.521066 | 0.269347 | -0.580413 | -0.564857 |
| PC-1 | -0.377418 | -0.923296 | -0.024492 | -0.066942 |
| PC-2 | 0.719566 | -0.244382 | -0.142126 | -0.634273 |
| PC-3 | 0.261286 | -0.123510 | -0.801449 | 0.523597 |