20. Optimizing a Function
In this notebook, we will explore how to find the optimal parameters of some function.
20.1. Data
Let’s simulate data.
\(z = f(x, y) = \sin \left( \sqrt{x^2 + y^2} \right)\)
[1]:
import pandas as pd
import numpy as np
x = np.arange(-5, 5, 0.25)
y = np.arange(-5, 5, 0.25)
x, y = np.meshgrid(x, y)
z = np.sin(np.sqrt(x**2 + y**2))
df = pd.DataFrame([(_x, _y, _z) for _x, _y, _z in zip(np.ravel(x), np.ravel(y), np.ravel(z))],
columns=['x', 'y', 'z'])
x.shape, y.shape, z.shape, df.shape
[1]:
((40, 40), (40, 40), (40, 40), (1600, 3))
20.2. Surface plot, Matplotlib
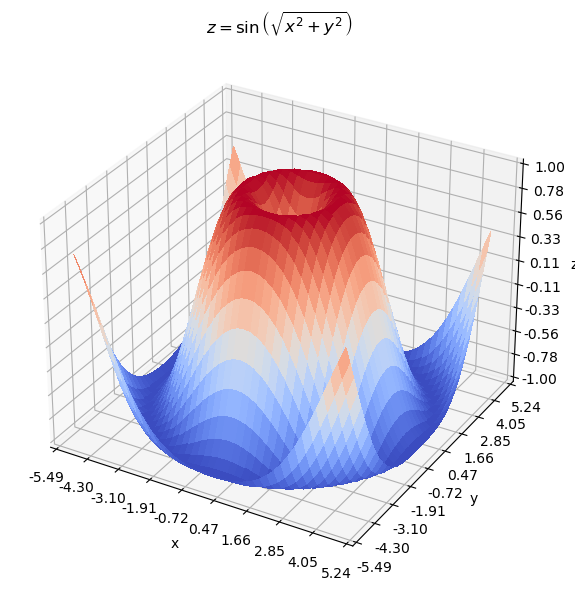
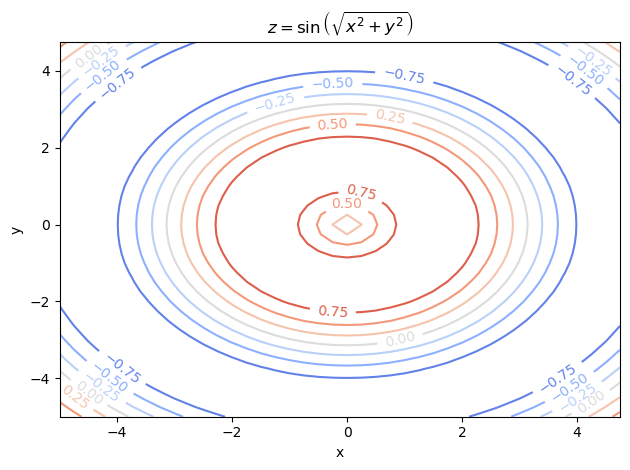
As you can see from the surface and contour plots below, the optimal parameters \(\theta = (x, y)\) should produce \(z \sim 1\) and is near the center of the graph (but not actually the center).
[2]:
import matplotlib.pyplot as plt
from matplotlib import cm
from matplotlib.ticker import LinearLocator
import numpy as np
fig, ax = plt.subplots(
subplot_kw={'projection': '3d'},
figsize=(6, 6)
)
surf = ax.plot_surface(x, y, z, cmap=cm.coolwarm, linewidth=0, antialiased=False)
ax.set_xlabel('x')
ax.set_ylabel('y')
ax.set_zlabel('z')
ax.xaxis.set_major_locator(LinearLocator(10))
ax.yaxis.set_major_locator(LinearLocator(10))
ax.zaxis.set_major_locator(LinearLocator(10))
ax.xaxis.set_major_formatter('{x:.02f}')
ax.yaxis.set_major_formatter('{x:.02f}')
ax.zaxis.set_major_formatter('{x:.02f}')
ax.set_title(r'$z = \sin \left( \sqrt{x^2 + y^2} \right)$')
fig.tight_layout()
[3]:
fig, ax = plt.subplots()
qcs = ax.contour(x, y, z, cmap=cm.coolwarm)
ax.clabel(qcs, inline=1, fontsize=10)
ax.set_title(r'$z = \sin \left( \sqrt{x^2 + y^2} \right)$')
ax.set_xlabel('x')
ax.set_ylabel('y')
fig.tight_layout()
20.3. Surface plot, Plotly
The static Matplotlib surface plot makes it hard to inspect the graph. A surface plot using Plotly is better since you can rotate the graph and inspect it.
[4]:
import plotly.graph_objects as go
fig = go.Figure(data=[go.Surface(
x=x,
y=y,
z=z,
colorscale='Viridis',
opacity=0.5,
)])
fig.update_layout(
title=r'$z = \sin \left( \sqrt{x^2 + y^2} \right)$',
autosize=False,
width=600,
height=600,
margin={'l': 65, 'r': 50, 'b': 65, 't': 90}
)
fig.update_xaxes(title='x')
fig.update_yaxes(title='y')
fig.show()
[5]:
fig = go.Figure(data =
go.Contour(
z=z,
x=df['x'].unique(),
y=df['y'].unique(),
colorscale='Viridis'
))
fig.update_layout(
title=r'$z = \sin \left( \sqrt{x^2 + y^2} \right)$',
autosize=False,
width=600,
height=600,
margin={'l': 65, 'r': 50, 'b': 65, 't': 90}
)
fig.show()
20.4. Optimal points
Since we simulated the data, we obviously know which points produces the optimal \(z\) values. There are several of them.
[6]:
df[df['z']==df['z'].max()]
[6]:
| x | y | z | |
|---|---|---|---|
| 578 | -0.5 | -1.5 | 0.999947 |
| 582 | 0.5 | -1.5 | 0.999947 |
| 734 | -1.5 | -0.5 | 0.999947 |
| 746 | 1.5 | -0.5 | 0.999947 |
| 894 | -1.5 | 0.5 | 0.999947 |
| 906 | 1.5 | 0.5 | 0.999947 |
| 1058 | -0.5 | 1.5 | 0.999947 |
| 1062 | 0.5 | 1.5 | 0.999947 |
20.5. Modeling
We can use a random forest model to approximate the function.
[7]:
from sklearn.ensemble import RandomForestRegressor
m = RandomForestRegressor(random_state=37, n_jobs=-1, n_estimators=50)
m.fit(df[['x', 'y']], df['z'])
[7]:
RandomForestRegressor(n_estimators=50, n_jobs=-1, random_state=37)
The non-validated error is pretty good.
[8]:
from sklearn.metrics import mean_absolute_error
mean_absolute_error(df['z'], m.predict(df[['x', 'y']]))
[8]:
0.011997664199503578
If we visulize the surface plot using the predicted \(\hat{z}\), you can see we have done a great job recovering the original surface plot.
[9]:
_x, _y = np.meshgrid(np.arange(-5, 5, 0.25), np.arange(-5, 5, 0.25))
_z = m.predict(pd.DataFrame([(_x_val, _y_val) for _x_val, _y_val in zip(np.ravel(_x), np.ravel(_y))], columns=['x', 'y'])).reshape(_x.shape)
_x.shape, _y.shape, _z.shape
[9]:
((40, 40), (40, 40), (40, 40))
[10]:
fig = go.Figure(data=[go.Surface(
x=_x,
y=_y,
z=_z,
colorscale='Viridis',
opacity=0.5,
)])
fig.update_layout(
title=r'$\hat{z} = \sin \left( \sqrt{x^2 + y^2} \right)$',
autosize=False,
width=600,
height=600,
margin={'l': 65, 'r': 50, 'b': 65, 't': 90}
)
fig.update_xaxes(title='x')
fig.update_yaxes(title='y')
fig.show()
20.6. Optimizing, Optuna
To find the optimal parameters, we can use Optuna to search for the best parameters.
[11]:
import optuna
optuna.logging.set_verbosity(optuna.logging.WARNING)
def objective(trial, m):
x = trial.suggest_float('x', -4, 4)
y = trial.suggest_float('y', -4, 4)
_df = pd.DataFrame([[x, y]], columns=['x', 'y'])
z = m.predict(_df)
return z[0]
study = optuna.create_study(**{
'study_name': 'opt-study',
'storage': 'sqlite:///_temp/opt-study.db',
'load_if_exists': True,
'direction': 'maximize',
'sampler': optuna.samplers.TPESampler(seed=37),
'pruner': optuna.pruners.MedianPruner(n_warmup_steps=10)
})
study.optimize(**{
'func': lambda t: objective(t, m),
'n_trials': 200,
'n_jobs': 6,
'show_progress_bar': True
})
/opt/anaconda3/lib/python3.9/site-packages/optuna/progress_bar.py:49: ExperimentalWarning:
Progress bar is experimental (supported from v1.2.0). The interface can change in the future.
Optuna does a decent job of finding the near-optimal parameters.
[12]:
study.best_params
[12]:
{'x': 1.555609216974581, 'y': -0.2773733254268651}
[13]:
study.best_value
[13]:
0.9971038658796584
20.7. Optimizing, Scipy
We can also find the optimal parameters using Scipy.
[14]:
import scipy.optimize as optimize
def f(tup):
x, y = tup
z = np.sin(np.sqrt(x ** 2 + y ** 2))
return -z
p_opt = optimize.fmin(f, np.array([0, 0]), ftol=1e-30, xtol=1e-30, maxiter=10_000, maxfun=10_000)
p_opt
Optimization terminated successfully.
Current function value: -1.000000
Iterations: 97
Function evaluations: 266
[14]:
array([1.54456722, 0.28585516])
The parameters found from Scipy is even more optimal than the ones we get from the data or Optuna!
[20]:
pd.Series([
-f(p_opt),
-f(np.array([0.5, 1.5])),
-f(np.array([study.best_params['x'], study.best_params['y']]))
], ['scipy', 'data', 'optuna'])
[20]:
scipy 1.000000
data 0.999947
optuna 0.999956
dtype: float64
[ ]: