8. K-means, BIC, AIC
8.1. Data
[1]:
import numpy as np
import random
from numpy.random import normal
from collections import namedtuple
Data = namedtuple('Data', 'X y')
random.seed(37)
np.random.seed(37)
def get_data(means, variances, labels, N=1000):
def get_X(sample_means, sample_variances, N):
return np.hstack([normal(m, v, N).reshape(-1, 1) for m, v in zip(sample_means, sample_variances)])
def get_y(label, N):
return np.full(N, label, dtype=np.int)
X = np.vstack([get_X(m, v, N) for m, v in zip(means, variances)])
y = np.hstack([get_y(label, N) for label in labels])
return Data(X, y)
# training
means = [
[5.0, 5.0],
[99.0, 99.0],
[15.0, 20.0]
]
variances = [
[1.0, 1.0],
[1.0, 1.0],
[1.0, 1.0]
]
labels = range(len(means))
X, y = get_data(means=means, variances=variances, labels=labels)
print(f'X shape = {X.shape}, y shape {y.shape}')
X shape = (3000, 2), y shape (3000,)
8.2. BIC, AIC and more
[2]:
from sklearn.cluster import KMeans
from sklearn.metrics import silhouette_score, davies_bouldin_score
from sklearn.metrics import homogeneity_score, completeness_score, v_measure_score
from sklearn.metrics import calinski_harabasz_score
from sklearn.mixture import GaussianMixture
from scipy.stats import multivariate_normal as mvn
def get_km(k, X):
km = KMeans(n_clusters=k, random_state=37)
km.fit(X)
return km
def get_bic_aic(k, X):
gmm = GaussianMixture(n_components=k, init_params='kmeans')
gmm.fit(X)
return gmm.bic(X), gmm.aic(X)
def get_score(k, X, y):
km = get_km(k, X)
y_pred = km.predict(X)
bic, aic = get_bic_aic(k, X)
sil = silhouette_score(X, y_pred)
db = davies_bouldin_score(X, y_pred)
hom = homogeneity_score(y, y_pred)
com = completeness_score(y, y_pred)
vms = v_measure_score(y, y_pred)
cal = calinski_harabasz_score(X, y_pred)
return k, bic, aic, sil, db, hom, com, vms, cal
The meaning of the scores.
BIClower is betterAIClower is bettersilhouettehigher is betterdavieslower is betterhomogeneityhigher is bettercompletenesshigher is betterv-measurehigher is bettercalinskihigher is better
[3]:
import pandas as pd
df = pd.DataFrame([get_score(k, X, y) for k in range(2, 11)],
columns=['k', 'BIC', 'AIC', 'silhouette',
'davies', 'homogeneity',
'completeness', 'vmeasure', 'calinski'])
df
[3]:
| k | BIC | AIC | silhouette | davies | homogeneity | completeness | vmeasure | calinski | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 2 | 29713.687099 | 29647.617056 | 0.941677 | 0.082989 | 0.57938 | 1.000000 | 0.733680 | 1.827224e+05 |
| 1 | 3 | 23709.628278 | 23607.520029 | 0.929479 | 0.099672 | 1.00000 | 1.000000 | 1.000000 | 2.623227e+06 |
| 2 | 4 | 23764.812594 | 23626.666140 | 0.731644 | 0.660094 | 1.00000 | 0.828906 | 0.906450 | 1.963947e+06 |
| 3 | 5 | 23812.883924 | 23638.699264 | 0.504188 | 1.032971 | 1.00000 | 0.703982 | 0.826279 | 1.672838e+06 |
| 4 | 6 | 23856.161363 | 23645.938498 | 0.306449 | 1.264110 | 1.00000 | 0.613262 | 0.760275 | 1.541983e+06 |
| 5 | 7 | 23898.831307 | 23652.570237 | 0.313627 | 1.128982 | 1.00000 | 0.573512 | 0.728958 | 1.437240e+06 |
| 6 | 8 | 23941.542123 | 23659.242848 | 0.324070 | 1.015535 | 1.00000 | 0.534216 | 0.696402 | 1.407921e+06 |
| 7 | 9 | 23986.361374 | 23668.023893 | 0.329671 | 0.952718 | 1.00000 | 0.502211 | 0.668629 | 1.422252e+06 |
| 8 | 10 | 24035.079248 | 23680.703562 | 0.326965 | 0.955671 | 1.00000 | 0.480658 | 0.649249 | 1.366897e+06 |
8.3. Visualize
[4]:
%matplotlib inline
import matplotlib.pyplot as plt
from sklearn.preprocessing import MinMaxScaler
plt.style.use('ggplot')
def plot_compare(df, y1, y2, x, fig, ax1):
ax1.plot(df[x], df[y1], color='tab:red')
ax1.set_title(f'{y1} and {y2}')
ax1.set_xlabel(x)
ax1.set_ylabel(y1, color='tab:red')
ax1.tick_params(axis='y', labelcolor='tab:red')
ax2 = ax1.twinx()
ax2.plot(df[x], df[y2], color='tab:blue')
ax2.set_ylabel(y2, color='tab:blue')
ax2.tick_params(axis='y', labelcolor='tab:blue')
def plot_contrast(df, y1, y2, x, fig, ax):
a = np.array(df[y1])
b = np.array(df[y2])
r_min, r_max = df[y1].min(), df[y1].max()
scaler = MinMaxScaler(feature_range=(r_min, r_max))
b = scaler.fit_transform(b.reshape(-1, 1))[:,0]
diff = np.abs(a - b)
ax.plot(df[x], diff)
ax.set_title('Scaled Absolute Difference')
ax.set_xlabel(x)
ax.set_ylabel('absolute difference')
def plot_result(df, y1, y2, x):
fig, axes = plt.subplots(1, 2, figsize=(10, 3))
plot_compare(df, y1, y2, x, fig, axes[0])
plot_contrast(df, y1, y2, x, fig, axes[1])
plt.tight_layout()
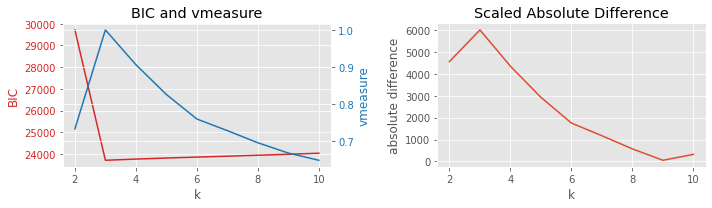
8.3.1. BIC vs silhouette
[5]:
plot_result(df, 'BIC', 'silhouette', 'k')
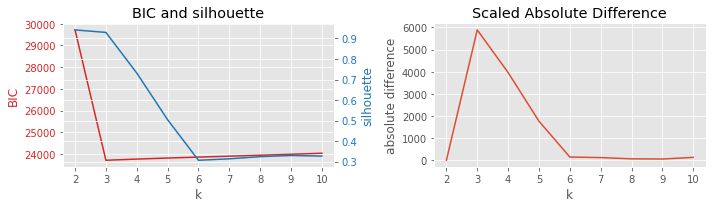
8.3.2. BIC vs Davies-Bouldin
[6]:
plot_result(df, 'BIC', 'davies', 'k')
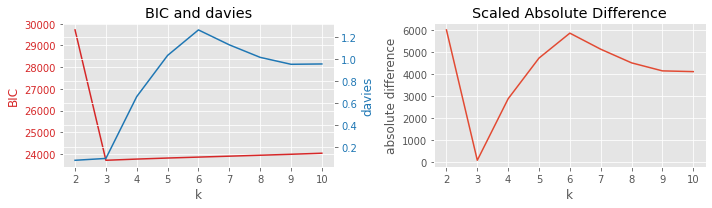
8.3.3. BIC vs homogeneity
[7]:
plot_result(df, 'BIC', 'homogeneity', 'k')
8.3.4. BIC vs completeness
[8]:
plot_result(df, 'BIC', 'completeness', 'k')
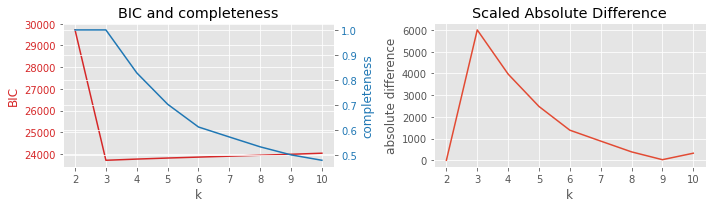
8.3.5. BIC vs Calinski-Harabasz
[9]:
plot_result(df, 'BIC', 'calinski', 'k')
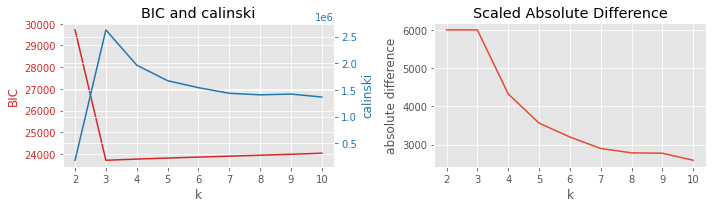
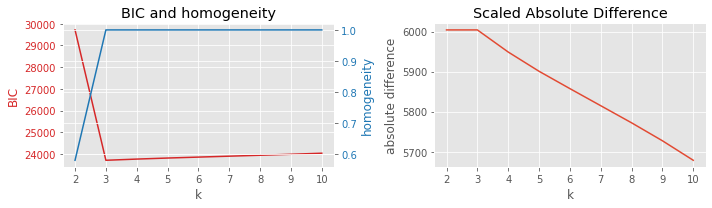
8.3.6. BIC vs v-measure
[10]:
plot_result(df, 'BIC', 'vmeasure', 'k')