13. Linear Mixed-Effect Regression
Linear Mixed-Effect Regression (LMER) take on the following functional form,
\(y = X \beta + Z u + \epsilon\),
where
\(y\) is the dependent variable
\(X\) is the design matrix for fixed effects (variables that have a constant effect on \(y\) regardless of groups),
\(Z\) is the design matrix for random effects (variables that have different effects on \(y\) across groups),
\(\beta\) is a vector of the coefficients for the fixed effects corresponding to \(X\),
\(u\) is a vector of the coefficients for the random effects corresponding to \(Z\), and
\(\epsilon\) is the random error.
LMER models are typically used to model data that can be grouped. For example, if you have a dataset of elementary students, they can be grouped into classrooms. The data being able to be grouped implies that the units (eg students) are related. Since the units are related, normal regression, like ordinary least-squares (OLS), may be inappropriate when they assume that units are independently and identically distributed (IID). Let’s take a look at LMER modeling.
13.1. Load data
The data and code below is taken from this post and were part of a study in which 30 female rats were randomly assigned to receive one of three doses (High, Low, or Control) of an experimental compound and the weights of their pups were measured as the effect of the chemical compound. The lowest unit of analysis is at the pup level, and these pups are further grouped into litters. We have the following variables.
\(y\) = weight
\(X\) = {sex, litsize, treatment}
\(Z\) = litter
[1]:
import pandas as pd
df = pd.read_csv('http://www-personal.umich.edu/~bwest/rat_pup.dat', sep='\t') \
.set_index(['pup_id'])
df.shape
[1]:
(322, 5)
[2]:
df.dtypes
[2]:
weight float64
sex object
litter int64
litsize int64
treatment object
dtype: object
[3]:
df.info()
<class 'pandas.core.frame.DataFrame'>
Int64Index: 322 entries, 1 to 322
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 weight 322 non-null float64
1 sex 322 non-null object
2 litter 322 non-null int64
3 litsize 322 non-null int64
4 treatment 322 non-null object
dtypes: float64(1), int64(2), object(2)
memory usage: 15.1+ KB
[4]:
df.isna().sum()
[4]:
weight 0
sex 0
litter 0
litsize 0
treatment 0
dtype: int64
[5]:
df.head()
[5]:
| weight | sex | litter | litsize | treatment | |
|---|---|---|---|---|---|
| pup_id | |||||
| 1 | 6.60 | Male | 1 | 12 | Control |
| 2 | 7.40 | Male | 1 | 12 | Control |
| 3 | 7.15 | Male | 1 | 12 | Control |
| 4 | 7.24 | Male | 1 | 12 | Control |
| 5 | 7.10 | Male | 1 | 12 | Control |
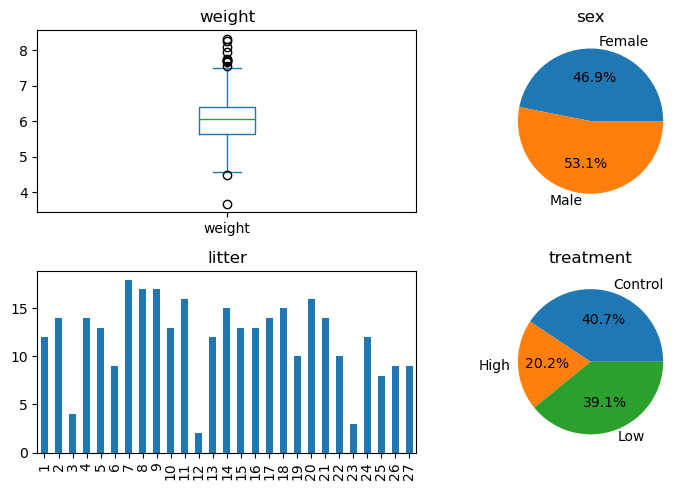
13.2. Visualize features
Now let’s pick some visualization to summarize each variable.
The median weight is 6.
There are more males (53%) than females (47%).
There is imbalance across the litters.
About 41% of the rat pups came from mothers that were not treated, followed by 39% whose mothers were treated with a low dose and 20% whose mothers were treated with a high dose.
[6]:
import matplotlib.pyplot as plt
import numpy as np
fig, ax = plt.subplots(2, 2, figsize=(8, 5))
ax = np.ravel(ax)
df['weight'].plot(kind='box', ax=ax[0])
df['sex'].value_counts().sort_index().plot(kind='pie', autopct='%1.1f%%', ax=ax[1])
df['litter'].value_counts().sort_index().plot(kind='bar', ax=ax[2])
df['treatment'].value_counts().sort_index().plot(kind='pie', autopct='%1.1f%%', ax=ax[3])
ax[0].set_title('weight')
ax[1].set_title('sex')
ax[2].set_title('litter')
ax[3].set_title('treatment')
ax[1].set_ylabel('')
ax[3].set_ylabel('')
fig.tight_layout()
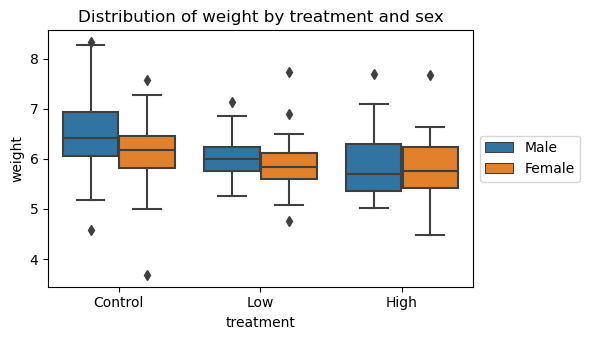
13.3. Visualize weights by treatment and sex
We can see that the median weight for both males and females trend from high to low as we move from the control to low to high-treatment groups.
[7]:
import seaborn as sns
fig, ax = plt.subplots(figsize=(6, 3.5))
sns.boxplot(data=df, x='treatment', y='weight', hue='sex', ax=ax)
ax.set_title('Distribution of weight by treatment and sex')
ax.legend(loc='center left', bbox_to_anchor=(1, 0.5))
fig.tight_layout()
13.4. Linear model
Let’s use a simple linear model for the data.
\(y = X \beta + \epsilon\)
[8]:
import statsmodels.formula.api as smf
ols_model = smf.ols(
formula='''weight ~ litsize + C(treatment) + C(sex, Treatment("Male"))''',
data=df) \
.fit()
ols_model.summary()
[8]:
| Dep. Variable: | weight | R-squared: | 0.409 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.401 |
| Method: | Least Squares | F-statistic: | 54.76 |
| Date: | Fri, 18 Aug 2023 | Prob (F-statistic): | 4.56e-35 |
| Time: | 09:45:07 | Log-Likelihood: | -231.84 |
| No. Observations: | 322 | AIC: | 473.7 |
| Df Residuals: | 317 | BIC: | 492.5 |
| Df Model: | 4 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 8.2240 | 0.156 | 52.864 | 0.000 | 7.918 | 8.530 |
| C(treatment)[T.High] | -0.9062 | 0.086 | -10.554 | 0.000 | -1.075 | -0.737 |
| C(treatment)[T.Low] | -0.4250 | 0.063 | -6.754 | 0.000 | -0.549 | -0.301 |
| C(sex, Treatment("Male"))[T.Female] | -0.2739 | 0.056 | -4.863 | 0.000 | -0.385 | -0.163 |
| litsize | -0.1249 | 0.010 | -12.236 | 0.000 | -0.145 | -0.105 |
| Omnibus: | 61.833 | Durbin-Watson: | 1.415 |
|---|---|---|---|
| Prob(Omnibus): | 0.000 | Jarque-Bera (JB): | 292.290 |
| Skew: | -0.693 | Prob(JB): | 3.39e-64 |
| Kurtosis: | 7.457 | Cond. No. | 82.0 |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
13.5. LMER, random intercept model
Now, we will use a LMER model allowing for the intercept to change. What do we mean by “allowing for the intercept to change?” We mean that there will be a different expected mean for each group (in the running example, each group is a litter of rat pups). Note the use of the groups parameter which is set to litter.
[9]:
rim_model = smf.mixedlm(
formula='''weight ~ litsize + C(treatment) + C(sex, Treatment('Male'))''',
data=df,
groups='litter') \
.fit()
rim_model.summary()
[9]:
| Model: | MixedLM | Dependent Variable: | weight |
| No. Observations: | 322 | Method: | REML |
| No. Groups: | 27 | Scale: | 0.1628 |
| Min. group size: | 2 | Log-Likelihood: | -198.4997 |
| Max. group size: | 18 | Converged: | Yes |
| Mean group size: | 11.9 |
| Coef. | Std.Err. | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 8.310 | 0.274 | 30.355 | 0.000 | 7.773 | 8.846 |
| C(treatment)[T.High] | -0.859 | 0.182 | -4.722 | 0.000 | -1.215 | -0.502 |
| C(treatment)[T.Low] | -0.429 | 0.150 | -2.849 | 0.004 | -0.723 | -0.134 |
| C(sex, Treatment('Male'))[T.Female] | -0.359 | 0.048 | -7.540 | 0.000 | -0.452 | -0.266 |
| litsize | -0.129 | 0.019 | -6.863 | 0.000 | -0.166 | -0.092 |
| litter Var | 0.097 | 0.085 |
13.6. LMER, random slope model, intercept and slopes independent
In this LMER model, we allow for the slope to change. In the LMER model where we allow for the intercept to change, the slopes are all the same and it is just the intercept that changes. Here, the slopes and intercepts changes per group, but the model does not assume interactions between the slopes and intercepts. Note the use of vc_formula parameter with the dictionary.
[10]:
rsm_model = smf.mixedlm(
formula='''weight ~ litsize + C(treatment) + C(sex, Treatment('Male'))''',
data=df,
groups='litter',
vc_formula = {'sex' : '0 + C(sex)'}) \
.fit()
rsm_model.summary()
[10]:
| Model: | MixedLM | Dependent Variable: | weight |
| No. Observations: | 322 | Method: | REML |
| No. Groups: | 27 | Scale: | 0.1565 |
| Min. group size: | 2 | Log-Likelihood: | -205.7020 |
| Max. group size: | 18 | Converged: | Yes |
| Mean group size: | 11.9 |
| Coef. | Std.Err. | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 8.231 | 0.234 | 35.185 | 0.000 | 7.772 | 8.689 |
| C(treatment)[T.High] | -0.839 | 0.144 | -5.827 | 0.000 | -1.121 | -0.557 |
| C(treatment)[T.Low] | -0.411 | 0.116 | -3.526 | 0.000 | -0.639 | -0.182 |
| C(sex, Treatment('Male'))[T.Female] | -0.356 | 0.102 | -3.508 | 0.000 | -0.555 | -0.157 |
| litsize | -0.124 | 0.016 | -7.915 | 0.000 | -0.155 | -0.093 |
| sex Var | 0.104 | 0.076 |
13.7. LMER, random slope model, intercept and slopes dependent
In this LMER model, we allow for the slopes and intercepts to change and assume an interaction between the two. Note the use of re_formula and string literal formula.
[11]:
ris_model = smf.mixedlm(
formula='''weight ~ litsize + C(treatment) + C(sex, Treatment('Male'))''',
data=df,
groups='litter',
re_formula = '1 + C(sex)') \
.fit()
ris_model.summary()
[11]:
| Model: | MixedLM | Dependent Variable: | weight |
| No. Observations: | 322 | Method: | REML |
| No. Groups: | 27 | Scale: | 0.1601 |
| Min. group size: | 2 | Log-Likelihood: | -196.3946 |
| Max. group size: | 18 | Converged: | Yes |
| Mean group size: | 11.9 |
| Coef. | Std.Err. | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 8.166 | 0.265 | 30.765 | 0.000 | 7.645 | 8.686 |
| C(treatment)[T.High] | -0.778 | 0.172 | -4.523 | 0.000 | -1.116 | -0.441 |
| C(treatment)[T.Low] | -0.387 | 0.137 | -2.821 | 0.005 | -0.656 | -0.118 |
| C(sex, Treatment('Male'))[T.Female] | -0.359 | 0.053 | -6.783 | 0.000 | -0.463 | -0.255 |
| litsize | -0.120 | 0.018 | -6.637 | 0.000 | -0.155 | -0.084 |
| litter Var | 0.063 | 0.074 | ||||
| litter x C(sex)[T.Male] Cov | 0.028 | 0.044 | ||||
| C(sex)[T.Male] Var | 0.013 | 0.062 |
Below is the estimated correlation coefficient between the random intercepts and random slopes. This correlation indicates that the litters with higher weight tend to have males that are of higher weight.
[12]:
ris_model.params['litter x C(sex)[T.Male] Cov'] / np.sqrt(ris_model.params['litter Var'] * ris_model.params['C(sex)[T.Male] Var'])
[12]:
0.9878805022022777
13.8. Performance
Let’s look at the non-validated performance measures. The MAE, RMSE, MAPE and \(R^2\) are all comparable between these models, but the log-likelihood is better for the random intercept, random slope model.
RIS: random intercept, random slope
RIM: random intercept
RSM: random slope
OLS: ordinary least square
[13]:
from sklearn.metrics import mean_absolute_error, mean_squared_error, mean_absolute_percentage_error, r2_score
def get_performance(m, X, y_true, Z=None, clusters=None):
if Z is not None and clusters is not None:
y_pred = m.predict(X, Z, clusters)
else:
y_pred = m.predict(X)
mae = mean_absolute_error(y_true, y_pred)
rmse = mean_squared_error(y_true, y_pred, squared=False)
mape = mean_absolute_percentage_error(y_true, y_pred)
rsq = r2_score(y_true, y_pred)
try:
ll = m.llf
except:
ll = np.nan
y_pred = pd.Series([mae, rmse, mape, rsq, ll], index=['mae', 'rmse', 'mape', 'r-sq', 'log-likelihood'])
return y_pred
lmer_df = pd.DataFrame([get_performance(m, df[['litsize', 'treatment', 'sex', 'litter']], df['weight'])
for m in [ols_model, rim_model, rsm_model, ris_model]],
index=['OLS', 'RIM', 'RSM', 'RIS']) \
.sort_values(['log-likelihood'], ascending=False)
lmer_df
[13]:
| mae | rmse | mape | r-sq | log-likelihood | |
|---|---|---|---|---|---|
| RIS | 0.383076 | 0.500760 | 0.064635 | 0.399894 | -196.394596 |
| RIM | 0.382219 | 0.499605 | 0.064361 | 0.402659 | -198.499691 |
| RSM | 0.381596 | 0.499295 | 0.064256 | 0.403400 | -205.702007 |
| OLS | 0.376124 | 0.497108 | 0.063282 | 0.408616 | -231.836723 |
13.9. Mixed-Effects Random Forest
Mixed-Effects Random Forest (MERF) is another approach to mixed-effect modeling. The functional form of the model is as follows,
\(y_i = f(X_i) + Z_i u_i + e_i\)
\(u_i \sim \mathcal{N}(0, D)\)
\(e_i \sim \mathcal{N}(0, \sigma_i)\)
where,
\(y_i\) is the vector of responses for the i-th cluster,
\(X_i\) is the fixed effects covariates associated with \(y_i\),
\(Z_i\) is the random effects covariates associated with the \(y_i\), and
\(e_i \sim \mathcal{N}(0, \sigma_i)\) is the vector of error for the i-th cluster.
The learned parameters in MERF are
\(f()\): a random forest that models the (potentially nonlinear) mapping from the fixed effect covariates to the response (common across all clusters),
\(D\): the covariance of the normal distribution from which each \(u_i\) is drawn, and
\(\sigma_i\): the variance of \(e_i\).
The random effects, \(Z_i u_i\), is still assumed to be linear.
To use MERF, install the following package.
pip install git+https://github.com/manifoldai/merf.git
Now let’s shape the data for input into MERF.
[14]:
sex_map = {'Male': 1, 'Female': 0}
treatment_map = {'Control': 0, 'Low': 1, 'High': 2}
X = df[['litsize', 'treatment', 'sex']] \
.assign(
sex=lambda d: d['sex'].map(sex_map),
treatment=lambda d: d['treatment'].map(treatment_map)
)
# Z = pd.DataFrame({'Z': np.ones(X.shape[0])}, index=X.index)
Z = df[['sex']].assign(sex=lambda d: d['sex'].map(sex_map))
clusters = df['litter']
y = df['weight']
X.shape, Z.shape, y.shape, clusters.shape
[14]:
((322, 3), (322, 1), (322,), (322,))
By default, MERF uses a random forest to model the fixed effects. You will see that the way the parameters are learned is through an iteration procedure. In particular, the parameters are learned using the Expectation-Maximization Algorithm.
[15]:
from merf import MERF
from sklearn.ensemble import RandomForestRegressor
merf = MERF(
fixed_effects_model=RandomForestRegressor(n_estimators=30, n_jobs=-1, random_state=37),
max_iterations=20
)
merf.fit(X, Z, clusters, y)
INFO [merf.py:307] Training GLL is -333.71605873307544 at iteration 1.
INFO [merf.py:307] Training GLL is -377.01192150920764 at iteration 2.
INFO [merf.py:307] Training GLL is -385.0144049452151 at iteration 3.
INFO [merf.py:307] Training GLL is -388.49413213816604 at iteration 4.
INFO [merf.py:307] Training GLL is -390.786958923229 at iteration 5.
INFO [merf.py:307] Training GLL is -392.6172680813943 at iteration 6.
INFO [merf.py:307] Training GLL is -394.1013482379355 at iteration 7.
INFO [merf.py:307] Training GLL is -395.3064926013952 at iteration 8.
INFO [merf.py:307] Training GLL is -396.2790567471805 at iteration 9.
INFO [merf.py:307] Training GLL is -397.0650753333675 at iteration 10.
INFO [merf.py:307] Training GLL is -397.70311641522136 at iteration 11.
INFO [merf.py:307] Training GLL is -398.2237484173181 at iteration 12.
INFO [merf.py:307] Training GLL is -398.65074861675413 at iteration 13.
INFO [merf.py:307] Training GLL is -399.00256401978334 at iteration 14.
INFO [merf.py:307] Training GLL is -399.29357896736184 at iteration 15.
INFO [merf.py:307] Training GLL is -399.5351033681913 at iteration 16.
INFO [merf.py:307] Training GLL is -399.7361114952454 at iteration 17.
INFO [merf.py:307] Training GLL is -399.90378716325665 at iteration 18.
INFO [merf.py:307] Training GLL is -400.0439266668877 at iteration 19.
INFO [merf.py:307] Training GLL is -400.16123920919347 at iteration 20.
[15]:
<merf.merf.MERF at 0x7f8f9830d850>
You can see from the merged results of LMER and MERF that MERF performs better (non-validated).
[16]:
merf_df = pd.DataFrame([get_performance(merf, X, y, Z, clusters)], index=['MERF'])
pd.concat([lmer_df, merf_df])
[16]:
| mae | rmse | mape | r-sq | log-likelihood | |
|---|---|---|---|---|---|
| RIS | 0.383076 | 0.500760 | 0.064635 | 0.399894 | -196.394596 |
| RIM | 0.382219 | 0.499605 | 0.064361 | 0.402659 | -198.499691 |
| RSM | 0.381596 | 0.499295 | 0.064256 | 0.403400 | -205.702007 |
| OLS | 0.376124 | 0.497108 | 0.063282 | 0.408616 | -231.836723 |
| MERF | 0.278688 | 0.376164 | 0.046600 | 0.661372 | NaN |
MERF is flexible enough to allow any linear or non-linear regressor to model the fixed effects. We are using a linear model here to model the fixed effects. This linear model for the fixed effects using the MERF API should reduce back to LMER.
[17]:
from sklearn.linear_model import LinearRegression
merf_linear = MERF(
fixed_effects_model=LinearRegression(),
max_iterations=20
)
merf_linear.fit(X, Z, clusters, y)
INFO [merf.py:307] Training GLL is -250.32226566191966 at iteration 1.
INFO [merf.py:307] Training GLL is -275.4248038416475 at iteration 2.
INFO [merf.py:307] Training GLL is -278.405863266045 at iteration 3.
INFO [merf.py:307] Training GLL is -279.14089177633406 at iteration 4.
INFO [merf.py:307] Training GLL is -279.39470078845005 at iteration 5.
INFO [merf.py:307] Training GLL is -279.50761907216946 at iteration 6.
INFO [merf.py:307] Training GLL is -279.56454572394097 at iteration 7.
INFO [merf.py:307] Training GLL is -279.59504914913975 at iteration 8.
INFO [merf.py:307] Training GLL is -279.6118859892217 at iteration 9.
INFO [merf.py:307] Training GLL is -279.62124285234916 at iteration 10.
INFO [merf.py:307] Training GLL is -279.6263549945002 at iteration 11.
INFO [merf.py:307] Training GLL is -279.62900945894233 at iteration 12.
INFO [merf.py:307] Training GLL is -279.6302331119333 at iteration 13.
INFO [merf.py:307] Training GLL is -279.63063176871344 at iteration 14.
INFO [merf.py:307] Training GLL is -279.6305682053091 at iteration 15.
INFO [merf.py:307] Training GLL is -279.63026066897373 at iteration 16.
INFO [merf.py:307] Training GLL is -279.62983973763113 at iteration 17.
INFO [merf.py:307] Training GLL is -279.62938219900144 at iteration 18.
INFO [merf.py:307] Training GLL is -279.62893170266136 at iteration 19.
INFO [merf.py:307] Training GLL is -279.6285115436138 at iteration 20.
[17]:
<merf.merf.MERF at 0x7f8fbc55deb0>
In this MERF example, we use something fancier, gradient boosting, to model the fixed effects.
[18]:
from lightgbm import LGBMRegressor
merf_lbgm = MERF(
fixed_effects_model=LGBMRegressor(),
max_iterations=20
)
merf_lbgm.fit(X, Z, clusters, y)
INFO [merf.py:307] Training GLL is -280.8403423506038 at iteration 1.
INFO [merf.py:307] Training GLL is -312.5902759696268 at iteration 2.
INFO [merf.py:307] Training GLL is -319.42122808679375 at iteration 3.
INFO [merf.py:307] Training GLL is -323.30449745760814 at iteration 4.
INFO [merf.py:307] Training GLL is -325.39602517001947 at iteration 5.
INFO [merf.py:307] Training GLL is -325.97589056606364 at iteration 6.
INFO [merf.py:307] Training GLL is -326.50014975589113 at iteration 7.
INFO [merf.py:307] Training GLL is -326.6869382890954 at iteration 8.
INFO [merf.py:307] Training GLL is -326.67740437411493 at iteration 9.
INFO [merf.py:307] Training GLL is -326.7714395331036 at iteration 10.
INFO [merf.py:307] Training GLL is -327.54464523883496 at iteration 11.
INFO [merf.py:307] Training GLL is -327.09840142944836 at iteration 12.
INFO [merf.py:307] Training GLL is -327.4145901178239 at iteration 13.
INFO [merf.py:307] Training GLL is -327.5149945679775 at iteration 14.
INFO [merf.py:307] Training GLL is -327.76484590606526 at iteration 15.
INFO [merf.py:307] Training GLL is -326.04470343017886 at iteration 16.
INFO [merf.py:307] Training GLL is -327.4639939165594 at iteration 17.
INFO [merf.py:307] Training GLL is -327.7250271749926 at iteration 18.
INFO [merf.py:307] Training GLL is -326.68773048443546 at iteration 19.
INFO [merf.py:307] Training GLL is -326.8288576814248 at iteration 20.
[18]:
<merf.merf.MERF at 0x7f8fbc54d370>
The table below compares the LMER and MERF models. In general, MERF models using a non-linear model for the fixed effects does better than LMER models across all (non-validated) performance measures. The problem with random forest and gradient boosting is they have a higher chance to overfit as they have higher variance, as opposed to the LMER models, which have higher bias (see bias-variance tradeoff).
[19]:
linear_df = pd.DataFrame([get_performance(merf_linear, X, y, Z, clusters)], index=['MERF_LINEAR'])
lgbm_df = pd.DataFrame([get_performance(merf_lbgm, X, y, Z, clusters)], index=['MERF_LGBM'])
pd.concat([lmer_df, merf_df, linear_df, lgbm_df])
[19]:
| mae | rmse | mape | r-sq | log-likelihood | |
|---|---|---|---|---|---|
| RIS | 0.383076 | 0.500760 | 0.064635 | 0.399894 | -196.394596 |
| RIM | 0.382219 | 0.499605 | 0.064361 | 0.402659 | -198.499691 |
| RSM | 0.381596 | 0.499295 | 0.064256 | 0.403400 | -205.702007 |
| OLS | 0.376124 | 0.497108 | 0.063282 | 0.408616 | -231.836723 |
| MERF | 0.278688 | 0.376164 | 0.046600 | 0.661372 | NaN |
| MERF_LINEAR | 0.303176 | 0.415935 | 0.051168 | 0.585982 | NaN |
| MERF_LGBM | 0.286231 | 0.405772 | 0.047979 | 0.605967 | NaN |
13.10. Learned parameters
Since model interpretability is important, you should look at the learned parameters. These are the parameters for the LMER model with random slopes and intercepts (slopes and intercepts are dependent).
[20]:
ris_model.params
[20]:
Intercept 8.165656
C(treatment)[T.High] -0.778370
C(treatment)[T.Low] -0.387161
C(sex, Treatment('Male'))[T.Female] -0.359242
litsize -0.119568
litter Var 0.395468
litter x C(sex)[T.Male] Cov 0.175758
C(sex)[T.Male] Var 0.080040
dtype: float64
For the MERF model using random forest to model the fixed effects, these are the feature importances of the fixed effects. You simply reference the random forest model and the feature_importances_ attribute. Litter size is apparently the most important feature for the fixed effects.
[21]:
pd.Series(merf.fe_model.feature_importances_, X.columns)
[21]:
litsize 0.558371
treatment 0.331581
sex 0.110048
dtype: float64
To look at the influence at the group level (litter), print out the trained_b. The nomenclature for MERF is to use \(b\) for the random effect coefficients (as opposed to LMER, which uses \(u\) for the random effect coefficients and \(b\) for the fixed effect coefficients). Do not let this cognitive friction confuse you.
[22]:
merf.trained_b.sort_index()
[22]:
| 0 | |
|---|---|
| 1 | -0.013010 |
| 2 | -0.003138 |
| 3 | 0.000726 |
| 4 | 0.002064 |
| 5 | 0.077942 |
| 6 | -0.002509 |
| 7 | -0.039926 |
| 8 | 0.211264 |
| 9 | -0.186452 |
| 10 | -0.147620 |
| 11 | -0.112385 |
| 12 | 0.000000 |
| 13 | -0.025229 |
| 14 | -0.234172 |
| 15 | 0.006974 |
| 16 | -0.011536 |
| 17 | -0.010374 |
| 18 | 0.171552 |
| 19 | 0.023750 |
| 20 | 0.109437 |
| 21 | 0.029966 |
| 22 | 0.045427 |
| 23 | 0.045619 |
| 24 | 0.008777 |
| 25 | 0.077531 |
| 26 | 0.053139 |
| 27 | -0.108956 |
13.11. Validation
Let’s split the data 70% (training) and 30% (testing) stratified by the litter identifier.
[60]:
from sklearn.model_selection import train_test_split
X_tr, X_te, y_tr, y_te = train_test_split(
df[['litsize', 'treatment', 'sex', 'litter']] \
.assign(
sex=lambda d: d['sex'].map(sex_map),
treatment=lambda d: d['treatment'].map(treatment_map)),
df['weight'],
test_size=0.30,
random_state=37,
shuffle=True,
stratify=df['litter']
)
Z_tr = X_tr[['sex']]
Z_te = X_te[['sex']]
C_tr = X_tr['litter']
C_te = X_te['litter']
X_tr = X_tr.drop(columns=['litter'])
X_te = X_te.drop(columns=['litter'])
[61]:
X_tr.shape, Z_tr.shape, C_tr.shape, y_tr.shape
[61]:
((225, 3), (225, 1), (225,), (225,))
[62]:
X_te.shape, Z_te.shape, C_te.shape, y_te.shape
[62]:
((97, 3), (97, 1), (97,), (97,))
Let’s make sure the proportions of litters in the training and testing folds are preserved.
[69]:
s1 = df['litter'].value_counts().sort_index()
s2 = C_tr.value_counts().sort_index()
s3 = C_te.value_counts().sort_index()
p1 = s1 / s1.sum()
p2 = s2 / s2.sum()
p3 = s3 / s3.sum()
pd.DataFrame([p1, p2, p3], index=['X', 'X_tr', 'X_te']).T
[69]:
| X | X_tr | X_te | |
|---|---|---|---|
| 1 | 0.037267 | 0.035556 | 0.041237 |
| 2 | 0.043478 | 0.044444 | 0.041237 |
| 3 | 0.012422 | 0.013333 | 0.010309 |
| 4 | 0.043478 | 0.044444 | 0.041237 |
| 5 | 0.040373 | 0.040000 | 0.041237 |
| 6 | 0.027950 | 0.026667 | 0.030928 |
| 7 | 0.055901 | 0.057778 | 0.051546 |
| 8 | 0.052795 | 0.053333 | 0.051546 |
| 9 | 0.052795 | 0.053333 | 0.051546 |
| 10 | 0.040373 | 0.040000 | 0.041237 |
| 11 | 0.049689 | 0.048889 | 0.051546 |
| 12 | 0.006211 | 0.004444 | 0.010309 |
| 13 | 0.037267 | 0.035556 | 0.041237 |
| 14 | 0.046584 | 0.048889 | 0.041237 |
| 15 | 0.040373 | 0.040000 | 0.041237 |
| 16 | 0.040373 | 0.040000 | 0.041237 |
| 17 | 0.043478 | 0.044444 | 0.041237 |
| 18 | 0.046584 | 0.048889 | 0.041237 |
| 19 | 0.031056 | 0.031111 | 0.030928 |
| 20 | 0.049689 | 0.048889 | 0.051546 |
| 21 | 0.043478 | 0.044444 | 0.041237 |
| 22 | 0.031056 | 0.031111 | 0.030928 |
| 23 | 0.009317 | 0.008889 | 0.010309 |
| 24 | 0.037267 | 0.035556 | 0.041237 |
| 25 | 0.024845 | 0.026667 | 0.020619 |
| 26 | 0.027950 | 0.026667 | 0.030928 |
| 27 | 0.027950 | 0.026667 | 0.030928 |
Now let’s validate.
[72]:
def get_model():
model = MERF(
fixed_effects_model=RandomForestRegressor(n_estimators=30, n_jobs=-1, random_state=37),
max_iterations=20
)
return model
def learn(X_tr, Z_tr, C_tr, y_tr):
model = get_model()
model.fit(X_tr, Z_tr, C_tr, y_tr)
return model
def get_results(X_tr, Z_tr, C_tr, y_tr, X_te, Z_te, C_te, y_te):
model = learn(X_tr, Z_tr, C_tr, y_tr)
perf_tr = get_performance(model, X_tr, y_tr, Z_tr, C_tr)
perf_te = get_performance(model, X_te, y_te, Z_te, C_te)
return pd.DataFrame([perf_tr, perf_te], index=['tr', 'te']).drop(columns=['log-likelihood']).T
get_results(X_tr, Z_tr, C_tr, y_tr, X_te, Z_te, C_te, y_te)
INFO [merf.py:307] Training GLL is -241.69649293327376 at iteration 1.
INFO [merf.py:307] Training GLL is -286.11740133632026 at iteration 2.
INFO [merf.py:307] Training GLL is -295.49781890130924 at iteration 3.
INFO [merf.py:307] Training GLL is -300.5394268800354 at iteration 4.
INFO [merf.py:307] Training GLL is -303.881513426677 at iteration 5.
INFO [merf.py:307] Training GLL is -306.11984582945183 at iteration 6.
INFO [merf.py:307] Training GLL is -307.9731073008684 at iteration 7.
INFO [merf.py:307] Training GLL is -309.2839553256728 at iteration 8.
INFO [merf.py:307] Training GLL is -310.34791724450554 at iteration 9.
INFO [merf.py:307] Training GLL is -311.1151258488934 at iteration 10.
INFO [merf.py:307] Training GLL is -311.7811932910758 at iteration 11.
INFO [merf.py:307] Training GLL is -312.2844557199853 at iteration 12.
INFO [merf.py:307] Training GLL is -312.67471304477255 at iteration 13.
INFO [merf.py:307] Training GLL is -312.93907731918256 at iteration 14.
INFO [merf.py:307] Training GLL is -313.26082011051926 at iteration 15.
INFO [merf.py:307] Training GLL is -313.4822415902106 at iteration 16.
INFO [merf.py:307] Training GLL is -313.66794283765677 at iteration 17.
INFO [merf.py:307] Training GLL is -313.8245804708934 at iteration 18.
INFO [merf.py:307] Training GLL is -313.95708548909306 at iteration 19.
INFO [merf.py:307] Training GLL is -314.11774594780064 at iteration 20.
[72]:
| tr | te | |
|---|---|---|
| mae | 0.265837 | 0.328698 |
| rmse | 0.366546 | 0.426194 |
| mape | 0.044403 | 0.054894 |
| r-sq | 0.681957 | 0.553932 |
The validated MERF performance is still pretty good.