9. Log-linear Models for Three-way Tables
Log-linear modeling is a type of analysis to understand the association between categorical variables. This approach uses regression to achieve its objective. However, the use of log-linear modeling is often multi-step, requires the production and comparison of many models and necessitates reflection on the tradeoff between goodness-of-fit GoF and model parsimony, degrees-of-freedom DoF. Log-linear modeling differs from logistic or multinomial regression in that the latter will typically
pick one of the categorical variables to be the dependent variable (conditional distribution of the dependent variable given the independent variables); in log-linear models, the dependent variable comes from the counts of the contingency table induced by the variables (joint distribution). As such, a very good way to understand log-linear modeling is to understand contingency tables.
In a three-way contingency table, we have 3 categorical variables, denoted as A, B and C and whose levels (values) are indexed as I, J and K, correspondingly. The log of the count \(\mu_{ijk} = n \pi_{ijk}\) (\(n\) is the sum of all the counts and \(\pi_{ijk} = \dfrac{n_{ijk}}{n}\)) is in a linear relationship to A, B and C in a finite number of ways, one of which is optimal and what we are trying to find using log-linear modeling. Here are some ways that
\(\mu_{ijk}\) may be related to A, B and C. (Remember that \(\mu_{ijk}\) is the product of the corresponding marginals divided by the total).
independence: A, B and C are all independent, denoted as
(A, B, C)\(\log{\mu_{ijk}} = \lambda + \lambda_i^A + \lambda_j^B + \lambda_k^C\)
joint independence: AB are jointly independent of C
(AB, C); AC are jointly independent of B(AC, B); BC are jointly independent of A(BC, A)\(\log{\mu_{ijk}} = \lambda + \lambda_i^A + \lambda_j^B + \lambda_k^C + \lambda_{jk}^{AB}\)
\(\log{\mu_{ijk}} = \lambda + \lambda_i^A + \lambda_j^B + \lambda_k^C + \lambda_{jk}^{AC}\)
\(\log{\mu_{ijk}} = \lambda + \lambda_i^A + \lambda_j^B + \lambda_k^C + \lambda_{jk}^{BC}\)
conditional independence: A and B are independent given C
(AC, BC); A and C are independent given B(AB, BC); B and C are independent give A(AB, AC)\(\log{\mu_{ijk}} = \lambda + \lambda_i^A + \lambda_j^B + \lambda_k^C + \lambda_{ik}^{AC} + \lambda_{jk}^{BC}\)
\(\log{\mu_{ijk}} = \lambda + \lambda_i^A + \lambda_j^B + \lambda_k^C + \lambda_{ij}^{AB} + \lambda_{jk}^{BC}\)
\(\log{\mu_{ijk}} = \lambda + \lambda_i^A + \lambda_j^B + \lambda_k^C + \lambda_{ij}^{AB} + \lambda_{ik}^{AC}\)
homogeneous association: there is no three way interaction
(AB, AC, BC)\(\log{\mu_{ijk}} = \lambda + \lambda_i^A + \lambda_j^B + \lambda_k^C + \lambda_{ij}^{AB} + \lambda_{ik}^{AC} + \lambda_{jk}^{BC}\)
saturated: there are main, pairwise and three-way effects
(ABC)\(\log{\mu_{ijk}} = \lambda + \lambda_i^A + \lambda_j^B + \lambda_k^C + \lambda_{ij}^{AB} + \lambda_{ik}^{AC} + \lambda_{jk}^{BC} + \lambda_{ijk}^{ABC}\)
The models above actually represent a Poisson regression model. Each cell count \(n_{ijk}\) is assumed to be Poisson distributed \(n_{ijk} \sim \operatorname{Pois}(\mu_{ijk})\). Thus, \(\lambda\) is the intercept and \(\lambda_i^A\), \(\lambda_j^B\), \(\lambda_k^C\) and so on are coefficients.
Each of these models will have a DoF and GoF associated with them. The GoF can be the log-likelihood of the model (higher is better) or the deviance \(G^2\) (lower is better). If we wanted to know the association between A, B and C, we can start from the independence model and move forward towards the saturated model (adding terms), or, we can start from the saturated model and move backward towards the independence model (remove terms). What is removed or added depends on how
the GoF and DoF changes; we must strike a balance between GoF and model parsimony. We typically compare models using a likelihood ratio test based on \(G^2\).
Lastly, the log-linear models above have graphical representation. Each log-linear model above asserts associations between the three variables.
9.1. Load data
This data is about UC-Berkely admissions from 1970. There are 3 categorical variables.
A: admissionB: sexC: department
[1]:
import pandas as pd
df = pd.read_csv('./data/berkeley-admission.csv')
df.shape
[1]:
(24, 4)
The three-way contingency table looks like the following.
[2]:
pd.crosstab(df['department'], [df['sex'], df['admission']], values=df['n'], aggfunc=lambda x: x)
[2]:
| sex | Female | Male | ||
|---|---|---|---|---|
| admission | Admitted | Rejected | Admitted | Rejected |
| department | ||||
| A | 89 | 19 | 512 | 313 |
| B | 17 | 8 | 353 | 207 |
| C | 202 | 391 | 120 | 205 |
| D | 131 | 244 | 139 | 279 |
| E | 94 | 299 | 53 | 138 |
| F | 24 | 317 | 22 | 351 |
We will use log-linear models to understand the relationship between these three variables.
9.2. Complete independence model (A, B, C)
\(\log{\mu_{ijk}} = \lambda + \lambda_i^A + \lambda_j^B + \lambda_k^C\)
[3]:
import networkx as nx
import matplotlib.pyplot as plt
plt.style.use('ggplot')
def draw_graph(g):
fig, ax = plt.subplots(figsize=(5, 5))
pos = {
'A': (200, 250),
'B': (150, 230),
'C': (250, 230)
}
params = {
'node_color': 'r',
'node_size': 350,
'node_shape': 's',
'alpha': 0.5,
'pos': pos,
'ax': ax
}
_ = nx.drawing.nx_pylab.draw_networkx_nodes(g, **params)
params = {
'font_size': 15,
'font_color': 'k',
'font_family': 'monospace',
'pos': pos,
'ax': ax
}
_ = nx.drawing.nx_pylab.draw_networkx_labels(g, **params)
params = {
'width': 1.5,
'alpha': 0.5,
'edge_color': 'b',
'arrowsize': 20,
'pos': pos,
'ax': ax
}
_ = nx.drawing.nx_pylab.draw_networkx_edges(g, **params)
g = nx.Graph()
g.add_node('A')
g.add_node('B')
g.add_node('C')
draw_graph(g)

[4]:
from patsy import dmatrices
import statsmodels.api as sm
dept_levels = ['F', 'A', 'B', 'C', 'D', 'E']
y, X = dmatrices('n ~ admission + sex + C(department, levels=dept_levels)', df, return_type='dataframe')
sm.GLM(y, X, family=sm.families.Poisson()).fit().summary()
[4]:
| Dep. Variable: | n | No. Observations: | 24 |
|---|---|---|---|
| Model: | GLM | Df Residuals: | 16 |
| Model Family: | Poisson | Df Model: | 7 |
| Link Function: | log | Scale: | 1.0000 |
| Method: | IRLS | Log-Likelihood: | -1128.1 |
| Date: | Wed, 09 Mar 2022 | Deviance: | 2097.1 |
| Time: | 12:13:30 | Pearson chi2: | 2.00e+03 |
| No. Iterations: | 5 | ||
| Covariance Type: | nonrobust |
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 4.7208 | 0.046 | 103.681 | 0.000 | 4.632 | 4.810 |
| admission[T.Rejected] | 0.4562 | 0.031 | 14.956 | 0.000 | 0.396 | 0.516 |
| sex[T.Male] | 0.3832 | 0.030 | 12.660 | 0.000 | 0.324 | 0.443 |
| C(department, levels=dept_levels)[T.A] | 0.2675 | 0.050 | 5.380 | 0.000 | 0.170 | 0.365 |
| C(department, levels=dept_levels)[T.B] | -0.1993 | 0.056 | -3.573 | 0.000 | -0.309 | -0.090 |
| C(department, levels=dept_levels)[T.C] | 0.2513 | 0.050 | 5.036 | 0.000 | 0.154 | 0.349 |
| C(department, levels=dept_levels)[T.D] | 0.1049 | 0.052 | 2.034 | 0.042 | 0.004 | 0.206 |
| C(department, levels=dept_levels)[T.E] | -0.2010 | 0.056 | -3.602 | 0.000 | -0.310 | -0.092 |
9.3. Joint independence
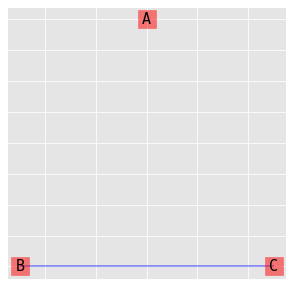
9.3.1. (A, BC)
\(\log{\mu_{ijk}} = \lambda + \lambda_i^A + \lambda_j^B + \lambda_k^C + \lambda_{jk}^{BC}\)
[5]:
g = nx.Graph()
g.add_node('A')
g.add_node('B')
g.add_node('C')
g.add_edge('B', 'C')
draw_graph(g)
[6]:
y, X = dmatrices('n ~ admission + sex + C(department, levels=dept_levels) + sex * C(department, levels=dept_levels)', df, return_type='dataframe')
sm.GLM(y, X, family=sm.families.Poisson()).fit().summary()
[6]:
| Dep. Variable: | n | No. Observations: | 24 |
|---|---|---|---|
| Model: | GLM | Df Residuals: | 11 |
| Model Family: | Poisson | Df Model: | 12 |
| Link Function: | log | Scale: | 1.0000 |
| Method: | IRLS | Log-Likelihood: | -517.91 |
| Date: | Wed, 09 Mar 2022 | Deviance: | 876.74 |
| Time: | 12:13:30 | Pearson chi2: | 797. |
| No. Iterations: | 7 | ||
| Covariance Type: | nonrobust |
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 4.8849 | 0.057 | 85.279 | 0.000 | 4.773 | 4.997 |
| admission[T.Rejected] | 0.4562 | 0.031 | 14.956 | 0.000 | 0.396 | 0.516 |
| sex[T.Male] | 0.0897 | 0.075 | 1.197 | 0.231 | -0.057 | 0.237 |
| C(department, levels=dept_levels)[T.A] | -1.1498 | 0.110 | -10.413 | 0.000 | -1.366 | -0.933 |
| C(department, levels=dept_levels)[T.B] | -2.6130 | 0.207 | -12.611 | 0.000 | -3.019 | -2.207 |
| C(department, levels=dept_levels)[T.C] | 0.5533 | 0.068 | 8.141 | 0.000 | 0.420 | 0.687 |
| C(department, levels=dept_levels)[T.D] | 0.0950 | 0.075 | 1.270 | 0.204 | -0.052 | 0.242 |
| C(department, levels=dept_levels)[T.E] | 0.1419 | 0.074 | 1.918 | 0.055 | -0.003 | 0.287 |
| sex[T.Male]:C(department, levels=dept_levels)[T.A] | 1.9436 | 0.127 | 15.325 | 0.000 | 1.695 | 2.192 |
| sex[T.Male]:C(department, levels=dept_levels)[T.B] | 3.0194 | 0.218 | 13.869 | 0.000 | 2.593 | 3.446 |
| sex[T.Male]:C(department, levels=dept_levels)[T.C] | -0.6911 | 0.102 | -6.784 | 0.000 | -0.891 | -0.491 |
| sex[T.Male]:C(department, levels=dept_levels)[T.D] | 0.0189 | 0.103 | 0.183 | 0.855 | -0.184 | 0.221 |
| sex[T.Male]:C(department, levels=dept_levels)[T.E] | -0.8112 | 0.116 | -7.010 | 0.000 | -1.038 | -0.584 |
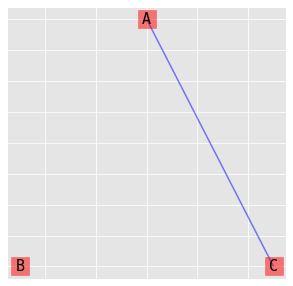
9.3.2. (B, AC)
\(\log{\mu_{ijk}} = \lambda + \lambda_i^A + \lambda_j^B + \lambda_k^C + \lambda_{ik}^{AC}\)
[7]:
g = nx.Graph()
g.add_node('A')
g.add_node('B')
g.add_node('C')
g.add_edge('A', 'C')
draw_graph(g)
[8]:
y, X = dmatrices('n ~ admission + sex + C(department, levels=dept_levels) + admission * C(department, levels=dept_levels)', df, return_type='dataframe')
sm.GLM(y, X, family=sm.families.Poisson()).fit().summary()
[8]:
| Dep. Variable: | n | No. Observations: | 24 |
|---|---|---|---|
| Model: | GLM | Df Residuals: | 11 |
| Model Family: | Poisson | Df Model: | 12 |
| Link Function: | log | Scale: | 1.0000 |
| Method: | IRLS | Log-Likelihood: | -700.56 |
| Date: | Wed, 09 Mar 2022 | Deviance: | 1242.1 |
| Time: | 12:13:30 | Pearson chi2: | 1.08e+03 |
| No. Iterations: | 6 | ||
| Covariance Type: | nonrobust |
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 2.9256 | 0.149 | 19.696 | 0.000 | 2.634 | 3.217 |
| admission[T.Rejected] | 2.6756 | 0.152 | 17.553 | 0.000 | 2.377 | 2.974 |
| sex[T.Male] | 0.3832 | 0.030 | 12.660 | 0.000 | 0.324 | 0.443 |
| C(department, levels=dept_levels)[T.A] | 2.5700 | 0.153 | 16.799 | 0.000 | 2.270 | 2.870 |
| C(department, levels=dept_levels)[T.B] | 2.0849 | 0.156 | 13.336 | 0.000 | 1.778 | 2.391 |
| C(department, levels=dept_levels)[T.C] | 1.9459 | 0.158 | 12.345 | 0.000 | 1.637 | 2.255 |
| C(department, levels=dept_levels)[T.D] | 1.7698 | 0.160 | 11.095 | 0.000 | 1.457 | 2.082 |
| C(department, levels=dept_levels)[T.E] | 1.1618 | 0.169 | 6.877 | 0.000 | 0.831 | 1.493 |
| admission[T.Rejected]:C(department, levels=dept_levels)[T.A] | -3.2691 | 0.167 | -19.567 | 0.000 | -3.597 | -2.942 |
| admission[T.Rejected]:C(department, levels=dept_levels)[T.B] | -3.2185 | 0.175 | -18.402 | 0.000 | -3.561 | -2.876 |
| admission[T.Rejected]:C(department, levels=dept_levels)[T.C] | -2.0600 | 0.167 | -12.306 | 0.000 | -2.388 | -1.732 |
| admission[T.Rejected]:C(department, levels=dept_levels)[T.D] | -2.0145 | 0.170 | -11.860 | 0.000 | -2.347 | -1.682 |
| admission[T.Rejected]:C(department, levels=dept_levels)[T.E] | -1.5861 | 0.180 | -8.822 | 0.000 | -1.939 | -1.234 |
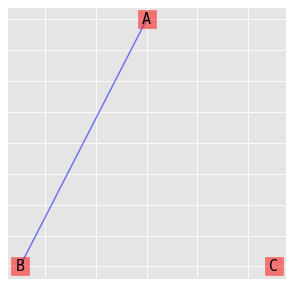
9.3.3. (C, AB)
\(\log{\mu_{ijk}} = \lambda + \lambda_i^A + \lambda_j^B + \lambda_k^C + \lambda_{ij}^{AB}\)
[9]:
g = nx.Graph()
g.add_node('A')
g.add_node('B')
g.add_node('C')
g.add_edge('A', 'B')
draw_graph(g)
[10]:
y, X = dmatrices('n ~ admission + sex + C(department, levels=dept_levels) + admission * sex', df, return_type='dataframe')
sm.GLM(y, X, family=sm.families.Poisson()).fit().summary()
[10]:
| Dep. Variable: | n | No. Observations: | 24 |
|---|---|---|---|
| Model: | GLM | Df Residuals: | 15 |
| Model Family: | Poisson | Df Model: | 8 |
| Link Function: | log | Scale: | 1.0000 |
| Method: | IRLS | Log-Likelihood: | -1081.2 |
| Date: | Wed, 09 Mar 2022 | Deviance: | 2003.4 |
| Time: | 12:13:30 | Pearson chi2: | 1.75e+03 |
| No. Iterations: | 5 | ||
| Covariance Type: | nonrobust |
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 4.4756 | 0.055 | 82.056 | 0.000 | 4.369 | 4.583 |
| admission[T.Rejected] | 0.8305 | 0.051 | 16.357 | 0.000 | 0.731 | 0.930 |
| sex[T.Male] | 0.7667 | 0.051 | 14.952 | 0.000 | 0.666 | 0.867 |
| C(department, levels=dept_levels)[T.A] | 0.2675 | 0.050 | 5.380 | 0.000 | 0.170 | 0.365 |
| C(department, levels=dept_levels)[T.B] | -0.1993 | 0.056 | -3.573 | 0.000 | -0.309 | -0.090 |
| C(department, levels=dept_levels)[T.C] | 0.2513 | 0.050 | 5.036 | 0.000 | 0.154 | 0.349 |
| C(department, levels=dept_levels)[T.D] | 0.1049 | 0.052 | 2.034 | 0.042 | 0.004 | 0.206 |
| C(department, levels=dept_levels)[T.E] | -0.2010 | 0.056 | -3.602 | 0.000 | -0.310 | -0.092 |
| admission[T.Rejected]:sex[T.Male] | -0.6112 | 0.064 | -9.567 | 0.000 | -0.736 | -0.486 |
9.4. Conditional independence
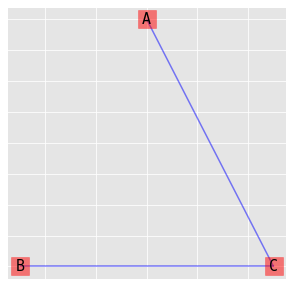
9.4.1. (AC, BC)
\(\log{\mu_{ijk}} = \lambda + \lambda_i^A + \lambda_j^B + \lambda_k^C + \lambda_{ik}^{AC} + \lambda_{jk}^{BC}\)
[11]:
g = nx.Graph()
g.add_node('A')
g.add_node('B')
g.add_node('C')
g.add_edge('A', 'C')
g.add_edge('B', 'C')
draw_graph(g)
[12]:
y, X = dmatrices('n ~ admission + sex + C(department, levels=dept_levels) + admission * C(department, levels=dept_levels) + sex * C(department, levels=dept_levels)', df, return_type='dataframe')
sm.GLM(y, X, family=sm.families.Poisson()).fit().summary()
[12]:
| Dep. Variable: | n | No. Observations: | 24 |
|---|---|---|---|
| Model: | GLM | Df Residuals: | 6 |
| Model Family: | Poisson | Df Model: | 17 |
| Link Function: | log | Scale: | 1.0000 |
| Method: | IRLS | Log-Likelihood: | -90.376 |
| Date: | Wed, 09 Mar 2022 | Deviance: | 21.686 |
| Time: | 12:13:30 | Pearson chi2: | 19.9 |
| No. Iterations: | 7 | ||
| Covariance Type: | nonrobust |
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 3.0896 | 0.153 | 20.253 | 0.000 | 2.791 | 3.389 |
| admission[T.Rejected] | 2.6756 | 0.152 | 17.553 | 0.000 | 2.377 | 2.974 |
| sex[T.Male] | 0.0897 | 0.075 | 1.197 | 0.231 | -0.057 | 0.237 |
| C(department, levels=dept_levels)[T.A] | 1.1527 | 0.182 | 6.334 | 0.000 | 0.796 | 1.509 |
| C(department, levels=dept_levels)[T.B] | -0.3289 | 0.254 | -1.297 | 0.195 | -0.826 | 0.168 |
| C(department, levels=dept_levels)[T.C] | 2.2479 | 0.164 | 13.687 | 0.000 | 1.926 | 2.570 |
| C(department, levels=dept_levels)[T.D] | 1.7599 | 0.168 | 10.447 | 0.000 | 1.430 | 2.090 |
| C(department, levels=dept_levels)[T.E] | 1.5047 | 0.176 | 8.559 | 0.000 | 1.160 | 1.849 |
| admission[T.Rejected]:C(department, levels=dept_levels)[T.A] | -3.2691 | 0.167 | -19.567 | 0.000 | -3.597 | -2.942 |
| admission[T.Rejected]:C(department, levels=dept_levels)[T.B] | -3.2185 | 0.175 | -18.402 | 0.000 | -3.561 | -2.876 |
| admission[T.Rejected]:C(department, levels=dept_levels)[T.C] | -2.0600 | 0.167 | -12.306 | 0.000 | -2.388 | -1.732 |
| admission[T.Rejected]:C(department, levels=dept_levels)[T.D] | -2.0145 | 0.170 | -11.860 | 0.000 | -2.347 | -1.682 |
| admission[T.Rejected]:C(department, levels=dept_levels)[T.E] | -1.5861 | 0.180 | -8.822 | 0.000 | -1.939 | -1.234 |
| sex[T.Male]:C(department, levels=dept_levels)[T.A] | 1.9436 | 0.127 | 15.325 | 0.000 | 1.695 | 2.192 |
| sex[T.Male]:C(department, levels=dept_levels)[T.B] | 3.0194 | 0.218 | 13.869 | 0.000 | 2.593 | 3.446 |
| sex[T.Male]:C(department, levels=dept_levels)[T.C] | -0.6911 | 0.102 | -6.784 | 0.000 | -0.891 | -0.491 |
| sex[T.Male]:C(department, levels=dept_levels)[T.D] | 0.0189 | 0.103 | 0.183 | 0.855 | -0.184 | 0.221 |
| sex[T.Male]:C(department, levels=dept_levels)[T.E] | -0.8112 | 0.116 | -7.010 | 0.000 | -1.038 | -0.584 |
9.4.2. (AB, BC)
\(\log{\mu_{ijk}} = \lambda + \lambda_i^A + \lambda_j^B + \lambda_k^C + \lambda_{ij}^{AB} + \lambda_{jk}^{BC}\)
[13]:
g = nx.Graph()
g.add_node('A')
g.add_node('B')
g.add_node('C')
g.add_edge('A', 'B')
g.add_edge('B', 'C')
draw_graph(g)
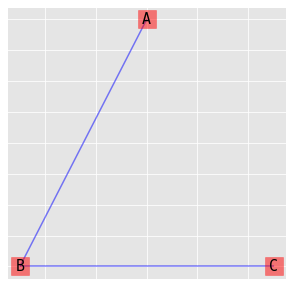
[14]:
y, X = dmatrices('n ~ admission + sex + C(department, levels=dept_levels) + admission * sex + sex * C(department, levels=dept_levels)', df, return_type='dataframe')
sm.GLM(y, X, family=sm.families.Poisson()).fit().summary()
[14]:
| Dep. Variable: | n | No. Observations: | 24 |
|---|---|---|---|
| Model: | GLM | Df Residuals: | 10 |
| Model Family: | Poisson | Df Model: | 13 |
| Link Function: | log | Scale: | 1.0000 |
| Method: | IRLS | Log-Likelihood: | -471.04 |
| Date: | Wed, 09 Mar 2022 | Deviance: | 783.02 |
| Time: | 12:13:31 | Pearson chi2: | 715. |
| No. Iterations: | 7 | ||
| Covariance Type: | nonrobust |
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 4.6396 | 0.065 | 71.737 | 0.000 | 4.513 | 4.766 |
| admission[T.Rejected] | 0.8305 | 0.051 | 16.357 | 0.000 | 0.731 | 0.930 |
| sex[T.Male] | 0.4731 | 0.086 | 5.528 | 0.000 | 0.305 | 0.641 |
| C(department, levels=dept_levels)[T.A] | -1.1498 | 0.110 | -10.413 | 0.000 | -1.366 | -0.933 |
| C(department, levels=dept_levels)[T.B] | -2.6130 | 0.207 | -12.611 | 0.000 | -3.019 | -2.207 |
| C(department, levels=dept_levels)[T.C] | 0.5533 | 0.068 | 8.141 | 0.000 | 0.420 | 0.687 |
| C(department, levels=dept_levels)[T.D] | 0.0950 | 0.075 | 1.270 | 0.204 | -0.052 | 0.242 |
| C(department, levels=dept_levels)[T.E] | 0.1419 | 0.074 | 1.918 | 0.055 | -0.003 | 0.287 |
| admission[T.Rejected]:sex[T.Male] | -0.6112 | 0.064 | -9.567 | 0.000 | -0.736 | -0.486 |
| sex[T.Male]:C(department, levels=dept_levels)[T.A] | 1.9436 | 0.127 | 15.325 | 0.000 | 1.695 | 2.192 |
| sex[T.Male]:C(department, levels=dept_levels)[T.B] | 3.0194 | 0.218 | 13.869 | 0.000 | 2.593 | 3.446 |
| sex[T.Male]:C(department, levels=dept_levels)[T.C] | -0.6911 | 0.102 | -6.784 | 0.000 | -0.891 | -0.491 |
| sex[T.Male]:C(department, levels=dept_levels)[T.D] | 0.0189 | 0.103 | 0.183 | 0.855 | -0.184 | 0.221 |
| sex[T.Male]:C(department, levels=dept_levels)[T.E] | -0.8112 | 0.116 | -7.010 | 0.000 | -1.038 | -0.584 |
9.4.3. (AB, AC)
\(\log{\mu_{ijk}} = \lambda + \lambda_i^A + \lambda_j^B + \lambda_k^C + \lambda_{ij}^{AB} + \lambda_{ik}^{AC}\)
[15]:
g = nx.Graph()
g.add_node('A')
g.add_node('B')
g.add_node('C')
g.add_edge('A', 'B')
g.add_edge('A', 'C')
draw_graph(g)
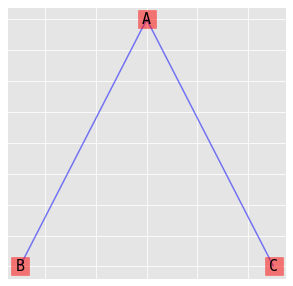
[16]:
y, X = dmatrices('n ~ admission + sex + C(department, levels=dept_levels) + admission * sex + admission * C(department, levels=dept_levels)', df, return_type='dataframe')
sm.GLM(y, X, family=sm.families.Poisson()).fit().summary()
[16]:
| Dep. Variable: | n | No. Observations: | 24 |
|---|---|---|---|
| Model: | GLM | Df Residuals: | 10 |
| Model Family: | Poisson | Df Model: | 13 |
| Link Function: | log | Scale: | 1.0000 |
| Method: | IRLS | Log-Likelihood: | -653.70 |
| Date: | Wed, 09 Mar 2022 | Deviance: | 1148.3 |
| Time: | 12:13:31 | Pearson chi2: | 1.02e+03 |
| No. Iterations: | 6 | ||
| Covariance Type: | nonrobust |
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 2.6804 | 0.152 | 17.688 | 0.000 | 2.383 | 2.977 |
| admission[T.Rejected] | 3.0500 | 0.158 | 19.335 | 0.000 | 2.741 | 3.359 |
| sex[T.Male] | 0.7667 | 0.051 | 14.952 | 0.000 | 0.666 | 0.867 |
| C(department, levels=dept_levels)[T.A] | 2.5700 | 0.153 | 16.799 | 0.000 | 2.270 | 2.870 |
| C(department, levels=dept_levels)[T.B] | 2.0849 | 0.156 | 13.336 | 0.000 | 1.778 | 2.391 |
| C(department, levels=dept_levels)[T.C] | 1.9459 | 0.158 | 12.345 | 0.000 | 1.637 | 2.255 |
| C(department, levels=dept_levels)[T.D] | 1.7698 | 0.160 | 11.095 | 0.000 | 1.457 | 2.082 |
| C(department, levels=dept_levels)[T.E] | 1.1618 | 0.169 | 6.877 | 0.000 | 0.831 | 1.493 |
| admission[T.Rejected]:sex[T.Male] | -0.6112 | 0.064 | -9.567 | 0.000 | -0.736 | -0.486 |
| admission[T.Rejected]:C(department, levels=dept_levels)[T.A] | -3.2691 | 0.167 | -19.567 | 0.000 | -3.597 | -2.942 |
| admission[T.Rejected]:C(department, levels=dept_levels)[T.B] | -3.2185 | 0.175 | -18.402 | 0.000 | -3.561 | -2.876 |
| admission[T.Rejected]:C(department, levels=dept_levels)[T.C] | -2.0600 | 0.167 | -12.306 | 0.000 | -2.388 | -1.732 |
| admission[T.Rejected]:C(department, levels=dept_levels)[T.D] | -2.0145 | 0.170 | -11.860 | 0.000 | -2.347 | -1.682 |
| admission[T.Rejected]:C(department, levels=dept_levels)[T.E] | -1.5861 | 0.180 | -8.822 | 0.000 | -1.939 | -1.234 |
9.5. Homogeneous model
\(\log{\mu_{ijk}} = \lambda + \lambda_i^A + \lambda_j^B + \lambda_k^C + \lambda_{ij}^{AB} + \lambda_{ik}^{AC} + \lambda_{jk}^{BC}\)
[17]:
g = nx.Graph()
g.add_node('A')
g.add_node('B')
g.add_node('C')
g.add_edge('A', 'B')
g.add_edge('A', 'C')
g.add_edge('B', 'C')
draw_graph(g)
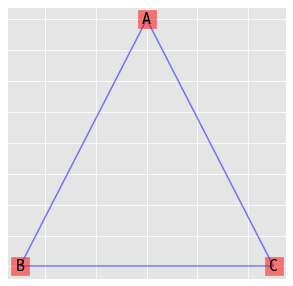
[18]:
y, X = dmatrices('n ~ (admission + sex + C(department, levels=dept_levels))**2', df, return_type='dataframe')
sm.GLM(y, X, family=sm.families.Poisson()).fit().summary()
[18]:
| Dep. Variable: | n | No. Observations: | 24 |
|---|---|---|---|
| Model: | GLM | Df Residuals: | 5 |
| Model Family: | Poisson | Df Model: | 18 |
| Link Function: | log | Scale: | 1.0000 |
| Method: | IRLS | Log-Likelihood: | -89.642 |
| Date: | Wed, 09 Mar 2022 | Deviance: | 20.217 |
| Time: | 12:13:31 | Pearson chi2: | 18.8 |
| No. Iterations: | 7 | ||
| Covariance Type: | nonrobust |
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 3.1364 | 0.157 | 20.011 | 0.000 | 2.829 | 3.444 |
| admission[T.Rejected] | 2.6256 | 0.158 | 16.646 | 0.000 | 2.316 | 2.935 |
| sex[T.Male] | -0.0018 | 0.106 | -0.017 | 0.987 | -0.210 | 0.207 |
| C(department, levels=dept_levels)[T.A] | 1.1359 | 0.182 | 6.242 | 0.000 | 0.779 | 1.493 |
| C(department, levels=dept_levels)[T.B] | -0.3422 | 0.253 | -1.351 | 0.177 | -0.839 | 0.154 |
| C(department, levels=dept_levels)[T.C] | 2.2233 | 0.165 | 13.484 | 0.000 | 1.900 | 2.546 |
| C(department, levels=dept_levels)[T.D] | 1.7466 | 0.168 | 10.387 | 0.000 | 1.417 | 2.076 |
| C(department, levels=dept_levels)[T.E] | 1.4814 | 0.176 | 8.407 | 0.000 | 1.136 | 1.827 |
| admission[T.Rejected]:sex[T.Male] | 0.0978 | 0.081 | 1.210 | 0.226 | -0.061 | 0.256 |
| admission[T.Rejected]:C(department, levels=dept_levels)[T.A] | -3.3057 | 0.170 | -19.448 | 0.000 | -3.639 | -2.973 |
| admission[T.Rejected]:C(department, levels=dept_levels)[T.B] | -3.2621 | 0.179 | -18.247 | 0.000 | -3.613 | -2.912 |
| admission[T.Rejected]:C(department, levels=dept_levels)[T.C] | -2.0442 | 0.168 | -12.177 | 0.000 | -2.373 | -1.715 |
| admission[T.Rejected]:C(department, levels=dept_levels)[T.D] | -2.0156 | 0.170 | -11.865 | 0.000 | -2.349 | -1.683 |
| admission[T.Rejected]:C(department, levels=dept_levels)[T.E] | -1.5676 | 0.180 | -8.688 | 0.000 | -1.921 | -1.214 |
| sex[T.Male]:C(department, levels=dept_levels)[T.A] | 2.0011 | 0.136 | 14.747 | 0.000 | 1.735 | 2.267 |
| sex[T.Male]:C(department, levels=dept_levels)[T.B] | 3.0759 | 0.223 | 13.802 | 0.000 | 2.639 | 3.513 |
| sex[T.Male]:C(department, levels=dept_levels)[T.C] | -0.6634 | 0.104 | -6.357 | 0.000 | -0.868 | -0.459 |
| sex[T.Male]:C(department, levels=dept_levels)[T.D] | 0.0459 | 0.106 | 0.434 | 0.664 | -0.161 | 0.253 |
| sex[T.Male]:C(department, levels=dept_levels)[T.E] | -0.7932 | 0.117 | -6.799 | 0.000 | -1.022 | -0.565 |
9.6. Saturated model
The saturated model is not estimated due to perfect separation.
\(\log{\mu_{ijk}} = \lambda + \lambda_i^A + \lambda_j^B + \lambda_k^C + \lambda_{ij}^{AB} + \lambda_{ik}^{AC} + \lambda_{jk}^{BC} + \lambda_{ijk}^{ABC}\)
9.7. Model Selection
With model selection, we can perform a likelihood ratio test between any two models.
[19]:
def get_deviance(m, f):
y, X = dmatrices(f, df, return_type='dataframe')
r = sm.GLM(y, X, family=sm.families.Poisson()).fit()
return {'model': m, 'df': r.df_resid, 'deviance': r.deviance}
formulas = {
'(A, B, C)': 'n ~ admission + sex + C(department, levels=dept_levels)',
'(A, BC)': 'n ~ admission + sex + C(department, levels=dept_levels) + sex * C(department, levels=dept_levels)',
'(B, AC)': 'n ~ admission + sex + C(department, levels=dept_levels) + admission * C(department, levels=dept_levels)',
'(C, AB)': 'n ~ admission + sex + C(department, levels=dept_levels) + admission * sex',
'(AC, BC)': 'n ~ admission + sex + C(department, levels=dept_levels) + admission * C(department, levels=dept_levels) + sex * C(department, levels=dept_levels)',
'(AB, BC)': 'n ~ admission + sex + C(department, levels=dept_levels) + admission * sex + sex * C(department, levels=dept_levels)',
'(AB, AC)': 'n ~ admission + sex + C(department, levels=dept_levels) + admission * sex + admission * C(department, levels=dept_levels)',
'(AB, AC, BC)': 'n ~ (admission + sex + C(department, levels=dept_levels))**2'
}
result_df = pd.DataFrame([get_deviance(m, f) for m, f in formulas.items()]).set_index('model')
result_df
[19]:
| df | deviance | |
|---|---|---|
| model | ||
| (A, B, C) | 16 | 2097.116655 |
| (A, BC) | 11 | 876.743972 |
| (B, AC) | 11 | 1242.058607 |
| (C, AB) | 15 | 2003.390913 |
| (AC, BC) | 6 | 21.685924 |
| (AB, BC) | 10 | 783.018230 |
| (AB, AC) | 10 | 1148.332865 |
| (AB, AC, BC) | 5 | 20.217417 |
Here are are comparing (AB, BC) \(H_0\) vs (AB, AC, BC) \(H_{\alpha}\). Since p < 0.01, we reject \(H_0\).
[20]:
from scipy.stats import chi2
chi_sq = result_df.loc['(AB, BC)'].deviance - result_df.loc['(AB, AC, BC)'].deviance
dof = result_df.loc['(AB, BC)'].df - result_df.loc['(AB, AC, BC)'].df
1 - chi2.cdf(chi_sq, dof)
[20]:
0.0
Here are are comparing (AC, BC) \(H_0\) vs (AB, AC, BC) \(H_{\alpha}\). Since p = 0.23, we fail to reject \(H_0\).
[21]:
chi_sq = result_df.loc['(AC, BC)'].deviance - result_df.loc['(AB, AC, BC)'].deviance
dof = result_df.loc['(AC, BC)'].df - result_df.loc['(AB, AC, BC)'].df
1 - chi2.cdf(chi_sq, dof)
[21]:
0.22558143086494165
What is the model (AC, BC) telling us? Remember, A is admission, B is sex and C is department. So, that we favor (AC, BC) as the best model, admission and sex are independent given department; meaning, there is no relationship between admission and sex given that we know which department they applied to, and the marginal association is spurious.