2. d-separation, Active Paths
Direction-dependent separation, or d-separation, is a rule for reading conditional independencies off a directed acyclic graph, DAG. The utility of d-separation is that it can determine if two mutually exclusive sets of variables, \(X\) and \(Y\), are independent give a third \(Z\). The output of d-separation is true when \(X\) and \(Y\) are independent given \(Z\), denoted as \(I(X,Y|Z)\), and false when
\(X\) and \(Y\) are dependent given \(Z\), denoted as \(D(X,Y|Z)\). If the output of d-separation is true, then \(X\) and \(Y\) are said to be d-separated given \(Z\); likewise, if the output of d-separation is false, then \(X\) and \(Y\) are said to be d-connected given \(Z\).
Let’s motivate the importance of d-separation. Let’s say we want to know if the treatment of an illness, \(T\), and the outcome of the illness, \(E\), are independent given race, \(R\). We want to know which of the following are true.
\(I(T,E|R)\)
\(D(T,E|R)\)
If \(T\) and \(E\) given \(R\) are d-separated, then once we know \(R\), \(T\) and \(E\) are no longer related. If \(T\) and \(E\) given \(R\) are d-connected, then when we know \(R\), \(T\) and \(E\) become related.
2.1. Graph example
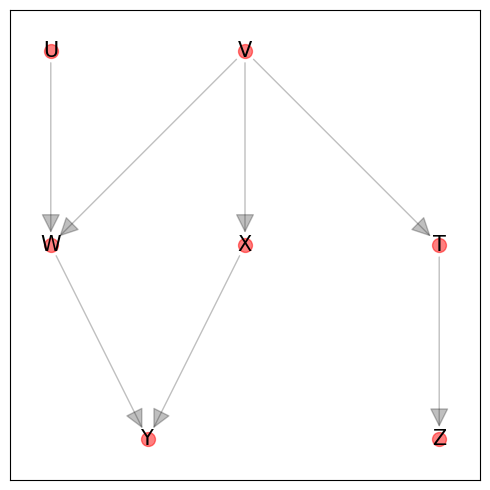
Here is a graph taken from this most excellent video tutorial on d-separation exercises. We will be using this graph to show some d-separation queries.
[1]:
import networkx as nx
from collections import namedtuple
Graph = namedtuple('Graph', 'u, d')
def get_edges(s):
edges = s.split('\n')
edges = map(lambda line: line.split('->'), edges)
edges = map(lambda tokens: tuple([t.strip() for t in tokens]), edges)
edges = list(edges)
return edges
def get_directed_graph(edges):
g = nx.DiGraph()
g.add_edges_from(edges)
return g
def get_undirected_graph(edges):
g = nx.Graph()
g.add_edges_from(edges)
return g
def get_graph(s):
e = get_edges(s)
d = get_directed_graph(e)
u = get_undirected_graph(e)
g = Graph(u, d)
return g
[2]:
s = '''
U -> W
V -> W
V -> X
V -> T
W -> Y
X -> Y
T -> Z
'''.strip()
g = get_graph(s)
[3]:
import matplotlib.pyplot as plt
pos = nx.nx_agraph.graphviz_layout(g.d, prog='dot')
fig, ax = plt.subplots(figsize=(5, 5))
nx.draw_networkx_nodes(
G=g.d,
pos=pos,
ax=ax,
node_size=100,
alpha=0.5,
node_color='red'
)
nx.draw_networkx_labels(
G=g.d,
pos=pos,
ax=ax,
font_size=15,
font_color='black',
alpha=1.0
)
nx.draw_networkx_edges(
G=g.d,
pos=pos,
alpha=0.25,
arrowsize=30,
ax=ax
)
fig.tight_layout()
2.2. Algorithm
Here’s an algorithm to determine if \(X\) and \(Y\) are d-separated given \(Z\).
Input:
\(G\): DAG
\(X\): start node
\(Y\): end node
\(Z\): evidence nodes
Output: True or False
Find all paths, \(P\), between \(X\) and \(Y\)
For each path \(p\) in \(P\)
if \(p\) is active, return False
return True
In the algorithm above, if we find any path in \(G\) between \(X\) and \(Y\) (given \(Z\)) that is active, then \(X\) and \(Y\) are d-connected; if we cannot find any active path between \(X\) and \(Y\) (given \(Z\)) in \(G\) then they are d-separated. The idea of an active and inactive path comes through understanding the configuration of triplets in a DAG. In general, there are 3 types of triplet configurations.
serial: \(X \rightarrow Z \rightarrow Y\) (causal chain)
diverging: \(X \leftarrow Z \rightarrow Y\) (common cause)
converging: \(X \rightarrow Z \leftarrow Y\) (common effect, also called a
v-structure)
For the serial and diverging configurations, without evidence of \(Z\), the configurations are active; if we do have evidence of \(Z\), then the serial and diverging configurations are inactive. The converging configuration is unique; without evidence of \(Z\), the configuration is inactive, and with evidence, the configuration is active. Active and inactive is about opening or blocking information, respectively.
A path between \(X\) and \(Y\) (given \(Z\)) is said to be active if all contiguous triplets are active (like a conjunctive boolean AND). If at least one of the contiguous triplets in the path is inactive, then the path is inactive (like a disjunctive boolean OR).
[4]:
def get_all_paths(g, start, stop):
return nx.all_simple_paths(g.u, start, stop)
def get_path_triplets(path):
if len(path) < 3:
return [path]
else:
return [path[i:i+3] for i in range(0, len(path) - 2)]
def get_triplet_type(g, x, z, y):
x_children = set(g.d.successors(x))
z_children = set(g.d.successors(z))
y_children = set(g.d.successors(y))
if z in x_children and y in z_children:
return 'serial'
if z in y_children and x in z_children:
return 'serial'
if x in z_children and y in z_children:
return 'diverging'
if z in x_children and z in y_children:
return 'converging'
raise Exception(f'cannot determine triplet configuration: x={x}, z={z}, y={y}')
def is_active_triplet(g, x, z, y, evidence=set()):
triplet_type = get_triplet_type(g, x, z, y)
if triplet_type in {'serial', 'diverging'}:
if z in evidence:
return False
else:
return True
else:
z_set = nx.descendants(g.d, z)
z_set.add(z)
if len(z_set & evidence) > 0:
return True
else:
return False
def is_active_path(g, path, evidence=set()):
if len(path) < 3:
if path[0] in evidence or path[-1] in evidence:
return False
else:
return True
path_triplets = get_path_triplets(path)
for x, z, y in path_triplets:
if not is_active_triplet(g, x, z, y, evidence):
return False
return True
def is_d_separated(g, x, y, evidence=set()):
paths = list(get_all_paths(g, x, y))
if len(paths) < 1:
return True
for path in get_all_paths(g, x, y):
if is_active_path(g, path, evidence):
return False
return True
2.3. Examples
Below are some exercise examples to query for d-separation based on the algorithm and code above.
2.3.1. Example 1
[5]:
is_d_separated(g, 'V', 'Z')
[5]:
False
2.3.2. Example 2
[6]:
is_d_separated(g, 'V', 'Z', {'T'})
[6]:
True
2.3.3. Example 3
[7]:
is_d_separated(g, 'U', 'V')
[7]:
True
2.3.4. Example 4
[8]:
is_d_separated(g, 'U', 'V', {'W'})
[8]:
False
2.3.5. Example 5
[9]:
is_d_separated(g, 'U', 'V', {'X'})
[9]:
True
2.3.6. Example 6
[10]:
is_d_separated(g, 'U', 'V', {'Y'})
[10]:
False
2.3.7. Example 7
[11]:
is_d_separated(g, 'U', 'V', {'Z'})
[11]:
True
2.3.8. Example 8
[12]:
is_d_separated(g, 'W', 'X')
[12]:
False
2.3.9. Example 9
[13]:
is_d_separated(g, 'X', 'T', {'V'})
[13]:
True
2.3.10. Example 10
[14]:
is_d_separated(g, 'X', 'W', {'U'})
[14]:
False
2.3.11. Example 11
[15]:
is_d_separated(g, 'Y', 'Z')
[15]:
False
2.3.12. Example 12
[16]:
is_d_separated(g, 'Y', 'Z', 'T')
[16]:
True
2.3.13. Example 13
[17]:
is_d_separated(g, 'Y', 'Z', {'X'})
[17]:
False
2.3.14. Example 14
[18]:
is_d_separated(g, 'Y', 'Z', {'V'})
[18]:
True
2.3.15. Example 15
[19]:
is_d_separated(g, 'W', 'Z', {'V'})
[19]:
True
2.3.16. Example 16
[20]:
is_d_separated(g, 'U', 'Z')
[20]:
True
2.3.17. Example 17
[21]:
is_d_separated(g, 'U', 'Z', {'Y'})
[21]:
False
2.3.18. Additional examples
[22]:
is_d_separated(g, 'U', 'W')
[22]:
False
[23]:
is_d_separated(g, 'U', 'W', {'U'})
[23]:
True
[24]:
is_d_separated(g, 'U', 'W', {'W'})
[24]:
True
[ ]: