6. Data Imputation with Autoencoders
Autoencoders may be used for data imputation. Let’s see how data imputation with autoencoder works.
6.1. Data
The data is sampled as follows.
\(X_0 \sim \mathcal{N}(0, 1)\)
\(X_1 \sim \mathcal{N}(1.1 + 4 X_0, 1)\)
\(X_2 \sim \mathcal{N}(2.3 - 0.5 X_0, 1)\)
[1]:
import numpy as np
import random
np.random.seed(37)
random.seed(37)
size = 1_000
X_0 = np.random.normal(0, 1, size=size)
X_1 = 1.1 + 4 * X_0 + np.random.normal(0, 1, size=size)
X_2 = 2.3 - 0.5 * X_0 + np.random.normal(0, 1, size=size)
X = np.hstack([X_0.reshape(-1, 1), X_1.reshape(-1, 1), X_2.reshape(-1, 1)])
X.shape
[1]:
(1000, 3)
6.2. Missing data
We will make 10% of the data missing randomly.
[2]:
import itertools
import pandas as pd
def make_missing(X, frac=0.1):
n = int(frac * X.shape[0] * X.shape[1])
rows = list(range(X.shape[0]))
cols = list(range(X.shape[1]))
coordinates = list(itertools.product(*[rows, cols]))
random.shuffle(coordinates)
coordinates = coordinates[:n]
M = np.copy(X)
for r, c in coordinates:
M[r, c] = np.nan
return pd.DataFrame(M, columns=[f'X_{i}' for i in range(X.shape[1])]), coordinates
df, coordinates = make_missing(X)
[3]:
df.isna().sum()
[3]:
X_0 99
X_1 102
X_2 99
dtype: int64
[4]:
df.isna().sum().sum()
[4]:
300
[5]:
import missingno as msno
_ = msno.matrix(df)
Denote the following.
N: represents data that is not missing (will be used for training)
T: represents data that is ground truth for missing data (will be used for validation)
M: represents data that is missing (will be used for testing)
[6]:
N_df = df.dropna()
T_df = pd.DataFrame(X[df.isnull().any(axis=1), :], columns=N_df.columns)
M_df = df[df.isnull().any(axis=1)]
N_df.shape, T_df.shape, M_df.shape
[6]:
((732, 3), (268, 3), (268, 3))
[7]:
T_df.iloc[0]
[7]:
X_0 -0.054464
X_1 0.222191
X_2 2.381753
Name: 0, dtype: float64
[8]:
M_df.iloc[0]
[8]:
X_0 -0.054464
X_1 NaN
X_2 2.381753
Name: 0, dtype: float64
6.3. Dataset, Data Loader
We will have to create our datasets and data loaders.
[9]:
import torch
from torch.utils.data import Dataset, DataLoader
from torchvision.transforms import *
class SampleDataset(Dataset):
def __init__(self, X, device, clazz=0):
self.__device = device
self.__clazz = clazz
self.__X = X
def __len__(self):
return self.__X.shape[0]
def __getitem__(self, idx):
item = self.__X[idx,:]
return item, self.__clazz
device = torch.device('cuda') if torch.cuda.is_available() else torch.device('cpu')
print(device)
N_ds = SampleDataset(X=N_df.values, device=device)
M_ds = SampleDataset(X=M_df.fillna(0.0).values, device=device)
N_dl = DataLoader(N_ds, batch_size=64, shuffle=True, num_workers=1)
M_dl = DataLoader(M_ds, batch_size=64, shuffle=True, num_workers=1)
cuda
6.4. Autoencoder
The two autoencoder architectures are adopted from the following.
[10]:
from torchvision import datasets
from torchvision import transforms
class AE1(torch.nn.Module):
def __init__(self, input_size):
super().__init__()
self.input_size = input_size
self.drop_out = torch.nn.Dropout(p=0.5)
self.encoder = torch.nn.Sequential(
torch.nn.Linear(input_size, 128),
torch.nn.ReLU(),
torch.nn.Linear(128, 64),
torch.nn.ReLU(),
torch.nn.Linear(64, 36),
torch.nn.ReLU(),
torch.nn.Linear(36, 18),
torch.nn.ReLU(),
torch.nn.Linear(18, 9)
)
self.decoder = torch.nn.Sequential(
torch.nn.Linear(9, 18),
torch.nn.ReLU(),
torch.nn.Linear(18, 36),
torch.nn.ReLU(),
torch.nn.Linear(36, 64),
torch.nn.ReLU(),
torch.nn.Linear(64, 128),
torch.nn.ReLU(),
torch.nn.Linear(128, input_size)
)
def forward(self, x):
drop_out = self.drop_out(x)
encoded = self.encoder(drop_out)
decoded = self.decoder(encoded)
return decoded
class AE2(torch.nn.Module):
def __init__(self, dim, theta=7):
super().__init__()
self.dim = dim
self.theta = theta
self.drop_out = torch.nn.Dropout(p=0.5)
self.encoder = torch.nn.Sequential(
torch.nn.Linear(dim+theta*0, dim+theta*1),
torch.nn.Tanh(),
torch.nn.Linear(dim+theta*1, dim+theta*2),
torch.nn.Tanh(),
torch.nn.Linear(dim+theta*2, dim+theta*3)
)
self.decoder = torch.nn.Sequential(
torch.nn.Linear(dim+theta*3, dim+theta*2),
torch.nn.Tanh(),
torch.nn.Linear(dim+theta*2, dim+theta*1),
torch.nn.Tanh(),
torch.nn.Linear(dim+theta*1, dim+theta*0)
)
def forward(self, x):
x = x.view(-1, self.dim)
x_missed = self.drop_out(x)
z = self.encoder(x_missed)
out = self.decoder(z)
out = out.view(-1, self.dim)
return out
6.5. Learning
We will train two autoencoder models and compare how they perform with data imputation.
[11]:
def train(model, optimizer):
loss_function = torch.nn.MSELoss()
epochs = 20
loss_df = []
for epoch in range(epochs):
losses = []
for (items, _) in N_dl:
items = items.to(device)
optimizer.zero_grad()
reconstructed = model(items)
loss = loss_function(reconstructed, items)
loss.backward()
optimizer.step()
losses.append(loss.detach().cpu().numpy().item())
losses = np.array(losses)
loss_df.append({
'epoch': epoch + 1,
'loss': losses.mean()
})
loss_df = pd.DataFrame(loss_df)
loss_df.index = loss_df['epoch']
loss_df = loss_df.drop(columns=['epoch'])
return loss_df
[12]:
model_1 = AE1(input_size=N_df.shape[1]).double().to(device)
opt_1 = torch.optim.Adam(model_1.parameters(), lr=1e-3, weight_decay=1e-8)
loss_1 = train(model_1, opt_1)
[13]:
model_2 = AE2(dim=N_df.shape[1]).double().to(device)
opt_2 = torch.optim.SGD(model_2.parameters(), momentum=0.99, lr=0.01, nesterov=True)
loss_2 = train(model_2, opt_2)
[14]:
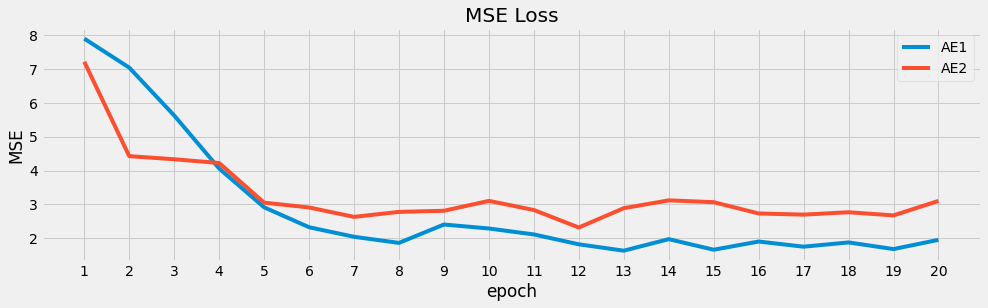
import matplotlib.pyplot as plt
plt.style.use('fivethirtyeight')
ax = loss_1['loss'].plot(kind='line', figsize=(15, 4), title='MSE Loss', ylabel='MSE', label='AE1')
_ = loss_2['loss'].plot(kind='line', figsize=(15, 4), title='MSE Loss', ylabel='MSE', ax=ax, label='AE2')
_ = ax.set_xticks(list(range(1, 21, 1)))
_ = ax.legend()
6.6. Imputation
Now we will impute the data using the two autoencoders. The performance will be the average L2 distance between the imputed and true data.
[15]:
def predict(m, items, device):
return m(items.to(device)).cpu().detach().numpy()
def get_imputation(m_v, p_v, t_v):
def get_value(m, p, t):
return p if pd.isna(m) else t
return np.array([get_value(m, p, t) for m, p, t in zip(m_v, p_v, t_v)])
def get_performance(model):
N_pred = np.vstack([predict(model, items, device) for items, _ in N_dl])
M_pred = np.vstack([predict(model, items, device) for items, _ in M_dl])
I_pred = np.array([get_imputation(M_df.values[r,:], M_pred[r,:], T_df.values[r,:]) for r in range(M_df.shape[0])])
n_perf = np.array([np.linalg.norm(N_df.values[r,:] - N_pred[r,:], 2) for r in range(N_df.shape[0])]).mean()
m_perf = np.array([np.linalg.norm(T_df.values[r,:] - I_pred[r,:], 2) for r in range(T_df.shape[0])]).mean()
return n_perf, m_perf
The first value is the training performance and the second value is the testing/validation performance. Lower is better. The results for the first autoencoder method is shown below.
[16]:
get_performance(model_1)
[16]:
(4.419058376740261, 2.0210263067003247)
The results for the second autoencoder method is shown below.
[17]:
get_performance(model_2)
[17]:
(4.414670724819407, 1.9455886899656865)